Journal of Animal Health and Production
Review Article
Comparative Diagnosis of Infectious Bacteria in Bovine Milk
Doaa Sedky1, Alaa A. Ghazy1, Nouran A. Soliman2, Raafat M. Shaapan3*
1Department of Parasitology and Animal Diseases, Veterinary Research Division, National Research Centre, Post Box 12622, El-Tahrir Street, Dokki, Giza, Egypt; 2Clinical Pharmacy Dep., Faculty of Pharmacy, University of Sadat City, Egypt; 3Department of Zoonotic Diseases, National Research Centre, Post Box 12622, El-Tahrir Street, Dokki, Giza, Egypt.
Abstract | Bovine milk is highly nutritive for people and calves as it contains macronutrients such as proteins, lipids and carbohydrates, in addition to micronutrients such as vitamins and minerals. Raw bovine milk is considered as a vehicle for many infectious bacterial diseases such as bovine tuberculosis, brucellosis, leptospirosis, and Mycoplasma infection. In addition to, infectious bacterial species associated with mastitis including Staphylococcus, Streptococcus, Corynebacteriun, Escherichia coli, Pseudomonas, Klebsiella, Clostridium perfringens and Salmonella species were also recorded. The aim of the present work is to high light on the diagnosis of the infectious bacteria in bovine milk. There are many available conventional and modern techniques used for the diagnosis of infectious bacteria in bovine milk. Conventional field tests such as somatic cell count, California mastitis test, electrical conductivity and pH tests have many advantages as they are convenient, non-expensive and can be used to monitor the udder health of the cattle. Microbial culture is a traditional technique which involves bacterial isolation, identification, microscopical examination, Gram staining, cultivation on enriched and selective media and studying of biochemical characters. Modern molecular diagnostic techniques are more valuable and offer more sensitive, fast and highly confident results such as polymerase chain reaction (PCR), multiplex PCR, real time PCR and whole genome sequencing. Another available diagnostic technique include immunological diagnosis using enzyme linked immunosorbent assays (ELISA) to detect not only antibody in bovine milk samples against many infectious bacteria, but also it is used to evaluate the levels of acute phase proteins such as hapatoglobin, c-reactive protein and mammary associated serum amyloid A3. The early and accurate diagnosis is necessary for achieving successful control programs against the infectious bacteria in bovine milk. It is recommended to use modern molecular diagnostic techniques as they are more valuable and offer more sensitive, fast and highly confident results, moreover they deliver a detailed understanding of the exact diagnosis. In addition, their ability to detect bacterial pathogens in dead or living stage from fresh or preserved milk samples was reported.
Keywords | Bovine milk, Bacteria, Comparative diagnosis, Foodborne pathogen, Techniques
Editor | Asghar Ali Kamboh, Sindh Agriculture University, Tandojam, Pakistan.
Received | April 24, 2020; Accepted | September 3, 2020; Published | September 25, 2020
*Correspondence | Raafat M. Shaapan, Department of Zoonotic Diseases, National Research Centre, Post Box 12622, El-Tahrir Street, Dokki, Giza, Egypt; Email: rmshaapan2005@yahoo.com
Citation | Sedky D, Ghazy AA, Soliman NA, Shaapan RM (2020). Comparative diagnosis of infectious bacteria in bovine milk. J. Anim. Health Prod. 8(4): 171-182.
DOI | http://dx.doi.org/10.17582/journal.jahp/2020/8.4.171.182
ISSN | 2308-2801
Copyright © 2020 Sedky et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Bovine milk is a source of nutrition for people and calf since it contains the supplements necessary for growth and progress of the calf. It could be a source of lipids, proteins, amino acids, minerals and vitamins. It contains immunoglobulins, hormones, growth factors, cytokines, nucleotides and peptides (Haug et al., 2007). Raw bovine milk is considered as a suitable growth medium for the multiplication of nearly all kinds of microorganisms (Kumar et al., 2010; Mahmoud et al., 2019). Bovine milk is considered as a vehicle for many infectious bacterial diseases. The most common infectious bacterial diseases that spread via bovine milk to young calves and human are bovine tuberculosis, brucellosis, mycoplasma infection, leptospirosis, and Q fever.
Bovine tuberculosis is a chronic infectious disease with economic and public health importance and it is of worldwide dissemination that influences animals (Leghari et al., 2020) and can be transmitted to humans by the ingestion of crude milk especially in the developing nations where bovine milk is ingested without prior pasteurization (Hassanain et al., 2009). This disease is caused by Mycobacterium bovis (M. bovis). Mycobacteria find their way out with respiratory tract secretions, milk, colostrum, feces, urine and, rarely, semen. Cattle-to-cattle transmission happens primarily by contact with aerosols or utilization of contaminated water and milk (Menzies and Neill, 2000). Chronic infection is characterized by development of huge caseous nodules, especially in the lungs (airborne disease) and intestinal tract (oral infection). In immunocompromised animals, early generalization may happen with spread of little nodules (granuloma) to organs, as well as the mammary gland (Domingo et al., 2014). Cattle with tuberculosis show diminished milk production, solidified masses within the udder, swelling of regional lymph nodes and watery milk (Ben et al., 2011). Tuberculous mastitis causes hardening of mammary quarters and swelling of supra mammary lymph nodes. Within the early stages of the infection, symptoms of clinical mastitis may have not seen in milk (subclinical mastitis). In chronic cases the milk contains fine clumps at the end of milking and the color may be altered to golden and Mycobacterium bovis is secreted in milk (Paes, 2010). Brucellosis is a highly infectious zoonotic bacterial disease which causes major reproductive losses in animals. Genus Brucella are Gram-negative, facultative intracellular pathogens which affect a wide range of mammals including humans, cattle, sheep, goats, pigs (Cutler et al., 2005). In spite of the performance of the National Brucellosis Control Program in Egypt from twenty two years ago, the infection is still endemic among ruminants and humans (Abdelbaset et al., 2018). Consumption of unpasteurized raw milk and dairy products made from this source could be a common source of infection to humans (Holt et al., 2011). In dairy cow, Brucella abortus replicate within the mammary gland and supra-mammary lymph nodes and persistently discharge the organism into milk. Brucella abortus spread through infected placentas, vaginal discharges and aborted fetuses (El-Mohammady et al., 2012). Mycoplasma bacterium belongs to the class Mollicutes which are characterized by their lack of cell wall, low guanine-cytosine content (G+C) content (23%–40%) and small size genome (Parte et al., 2011). Mycoplasma bovis is the most important etiological agent of different bovine diseases such as mastitis in cattle, pneumonia and arthritis in calves as well as genital disorders (Maunsell et al., 2011). Mycoplasma bovis infections tend to be chronic and responsible for significant economic losses in cattle and milk production (Eissa et al., 2012). The disease is characterized by abnormal secretion followed by serious mastitis which did not respond to treatment with antibiotics. Leptospirosis is an infectious zoonotic disease which happens by the genus Leptospira. Bovine leptospirosis causes serious economic losses as a result of abortion, stillbirths, infertility, mastitis, reduce in milk production and death. The disease commonly occurs in tropical and sub-tropical countries (Langoni et al., 1999). Milk drop syndrome happens in lactating dairy cows due to the multiplication of the organism in the lactating mammary gland. The disease is characterized by fever, anorexia, immobility, and agalactia. The decline in milk yield may remain up to eight weeks and milk production will return to normal within 10 to 14 days. The milk may have a high leukocyte count for 14 days. Few publications have recorded the occurrence of mastitis due to leptospirosis (Garoussi et al., 2017). Q fever is a zoonotic disease which affects humans and domestic animals. The causative agent is a Gram-negative intracellular bacterium and is known as Coxiella burnetii, the disease is transmitted mainly through inhaling aerosols and dust from contaminated materials shed by infected animals (Gwida et al., 2012).
A variety of Gram positive and Gram negative infectious bacteria were isolated from mastitic bovine milk. Gram-positive major pathogens such as Staphylococcus aureus (S. aureus), coagulase - negative staphylococci (CNS), Streptococcus dysgalactiae (S. dysgalactiae), Streptococcus agalactiae (S. agalactiae), Corynebacteriun pyogenes (C. pyogenes), and Streptococcus uberis (S. uberis). The Gram-negative major pathogen includes Escherichia coli (E. coli), Pseudomonas aeruginosa (P. aeruginosa), Klebsiella pneumonia (K. pneumonia) and Serratia species. On the other hand, the minor pathogens include Micrococcus species, Enterococcus faecalis (E. faecalis), Streptococcus pyogenes (S. pyogenes), Clostridium perfringens (C. perfringens), Salmonella species and other Enterobacteriaceae (Petzer et al., 2017). Mastitis is the infection that has the most significant effect on the milk quality. There is no noticeable difference between the milk produced by cows with subclinical mastitis and that produced by healthy cows. Nevertheless, milk from cows with clinical mastitis differs in appearance as it may contain flakes, clots, or blood or may acquire different colors (Islam et al., 2010). In Lower Egypt, out of 360 female buffaloes examined, 5.8% were reported as suffering from mastitis (Ghazy et al., 2007a).
There are many available conventional and modern techniques used for the diagnosis of infectious bacteria in bovine milk. Therefore, the aim of the present study is to highlight on the methods of diagnosis of the infectious bacteria in bovine milk.
Conventional diagnosis of bacteria in bovine milk
Conventional field tests
The somatic cell count (SCC) is an important indicator of milk quality. White blood cells constitute the majority of SCC (75%), while epithelia represent 25%. The number of SCC increases with the increase in immune response as in cases of infection with pathogenic bacteria (Harmon, 1994). The SCC in normal milk is less than 100,000 cells/ml, on the other hand in case of bacterial infections the SCC will increase to reach levels greater than 1000,000 cells/ml (Bytyqi et al., 2010). California mastitis test (CMT) is a simple cow-side assay used for diagnosis of the subclinical mastitis as this test indirectly measures the somatic cell count in bovine milk samples. The test is simply based on using a bromocresol-purple-containing detergent which is used to disturb the cell membrane of somatic cells, subsequently the released nucleic acid reacts with the reagent of the test and aggregates forming a gel-like matrix with a viscosity that is proportional to the leukocyte count (Olechnowicz and Jaśkowski, 2012). The reaction is scored on scale ranges from 0 to 3, where in 0 the mixture remains unchanged, while in 3 a solid gel is produced. Moreover, score 2 or 3 are considered as a positive result (Whyte et al., 2005). Other methods are described as Porta check, where this assay depends on an esterase-catalysed enzymatic reaction to measure the SCC in milk. Moreover, Fossomatic somatic cell counter operates on the principle of optical fluorescence, where ethidium bromide penetrates cell membrane of leukocyte and intercalates with nuclear DNA, and the resulting fluorescent signal is used as an indicator for the presence of the SCC in milk (Viguier et al., 2009). Bulk Tank Somatic Cell Count (BTSCC) is used as a routine monthly monitor for screening the milk quality. If BTSCC levels are lower than 2x105 cells mL-1 this points to low level of infection, on the other hand in clinical or chronic bacterial infections the BTSCC’s level will increase more than 5x105 cells mL-1 (Schepers et al., 1997). The sensitivity and specificity of the SCC test was calculated to detect Gram positive and Gram negative and minor pathogens in quarter and composite milk (combination of bovine milk sample from four udder quarters). They found that the percentage of Gram-positive pathogens was 88.9% in quarter, and 79.9% in composite samples, while the percentage of minor pathogens was 61.4% in quarter and 51.7% in composite samples. They added that the sensitivity of the SCC test to detect major pathogens was 88.2% in quarter samples and 84.2% in composite milk samples; on the other hand the specificity was 57.7% in quarter and 52.8% in composite milk samples (Petzer et al., 2017). Electrical conductivity and pH tests are convenient and non-expensive tests which can be used by non-specialized personnel on regular basis to monitor the udder health of the cattle (Pitkala et al., 2005; Viguier et al., 2009). Mast-O-Test is used for analysis of the electric conductivity of the milk for early detection of subclinical mastitis where it identifies the relation between the electric conductivity and the milk content of lactose and salt (Musser et al., 1998).
In vitro microbial culture
In vitro microbial culture is considered as fundamental diagnostic test for isolation of infectious bacteria. For specific detection of bacteria, a swab of fresh milk samples is cultivated in a specific media and then the isolates are subjected to microbiological and biochemical identification (Pyorala, 2003). Milk samples are cultivated in highly enriched media such as brain heart agar, blood agar base, mannitol salt agar, MacConkey agar, Mueller-Hinton agar, and Edward’s medium. Plates are incubated at 37°C for 24 and 48 hours. Colonies will be identified by their morphology and Gram-staining. Coagulase tests with rabbit plasma is used to differentiate between S. aureus and coagulase negative Staphylococcus (CNS) (Rajeev et al., 2009). Moreover, esculin hydrolysis and Christie-Atkins Munch-Petersen (CAMP) test are used to differentiate between Streptococcus agalactiae and other Streptococcus species. Gram-negative bacteria will be identified by sub culturing on differential and selective media and tested to oxidase activity, acid production, sugar fermentation test, indole test, Voges–Proskauer test (VP) and hydrogen sulfide production (National Mastitis Council, 1999) (Figure 1).
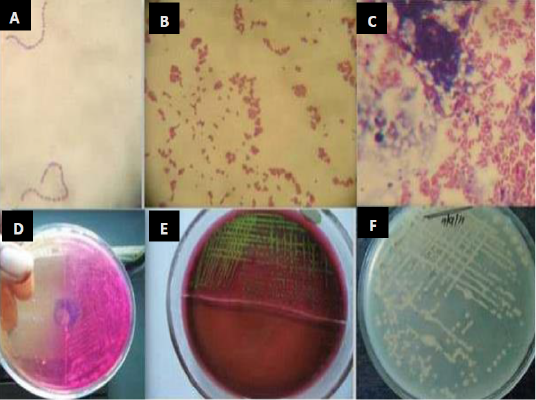
Figure 1: Isolation and identification of Staphylococcus spp., Streptococcus and E. coli from bovine mastitic milk sample. (A) Streptococcus spp. (B) Staphylococcus spp. (C) Gram negative bacilli (D) colony growth on mannitol salt agar (E) metallic sheen on EMB agar by E. coli isolates (F) colonies on muller hinton agar (Mohanty et al., 2013).
The main disadvantage of bacterial cultivation is that it requires a specific medium and takes a long time. Moreover, the culture is limited to the recognition of live cells and thus negative results need further investigation (Rajapaksha et al., 2019). The comparative diagnosis by use of mannitol fermentation assay (Mannitol salt agar; MSA), and deoxyribonuclease (DNase) assay for Staphylococcus aureus identification in subclinical bovine mastitis milk samples revealed 130 Staphylococci isolates by culturing on selective media, Gram-staining and catalase test. While 32 Staphylococcus aureus isolates were identified using the tube coagulase test (TCT), 102 isolates were identified by mannitol fermentation assay and 74 isolates were identified by deoxyribonuclease test (Pumipuntu et al., 2017). The sensitivity of MSA reached 100%, while the specificity reached 28.57%. On the other hand, the sensitivity of DNase reached 53.13%, while the specificity reached 41.84%. The specificity of deoxyribonuclease assay is higher than mannitol fermentation assay. However, the DNase assay showed 26.79% false negative results, while the MSA didn’t show any false-negative result. They concluded that the deoxyribonuclease assay can be used for diagnosis of Staphylococci aureus infection in bovine milk sample (Figure 2).
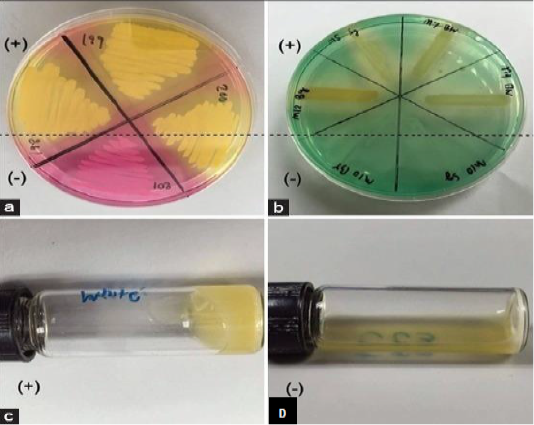
Figure 2: Screening tests for identification of Staphylococcus aureus from subclinical bovine mastitic milk sample. mannitol salt agar (a): deoxyribonuclease test (b): tube coagulase test positive result (c) and coagulase test negative result (d) (Pumipuntu et al., 2017).
Isolation and identification of Pseudomonas aeruginosa in milk samples from dairy farms in India using selective media cetrimide and pseudomonas agar was carried in an earlier study (Banerjee et al., 2017). 23 out of 352 (6.5%) samples yielded typical Pseudomonas spp., and 19 P. aeruginosa were isolated with percentage of 5.4% which were characterized by blue-green fluorescence due to presence of pyoverdin pigment detected under ultraviolet light (Banerjee et al., 2017) (Figure 3).
The isolation of Mycobacterium bovis needs enriched selective media as Lowenstein-Jensen medium which contains egg (Cheng et al., 1994). The green color of Lowenstein-Jensen medium is attributed to the presence of malachite green dye which prevents the growth of other bacteria. Also, Stone brink is another egg based enriched medium which contains pyruvate as a growth stimulant of M. bovis, the colonies of Mycobacterium bovis are yellowish and granular with a creamy appearance (Stonebrink, 1958). Bovine milk samples are incubated at 37 °C under aerobic conditions and assessed week after week up to ninety days. Phenotypic identification depends on isolation time, morphologic appearance of the colonies and pigment production (Figure 4). In a study at Giza province, Egypt M. bovis was isolated from milk samples of 19 out of 50 dairy cattle samples (38.0%) exposed to an outbreak of bovine tuberculosis using Lowenstein-Jensen medium (Ghazy et al, 2007b).
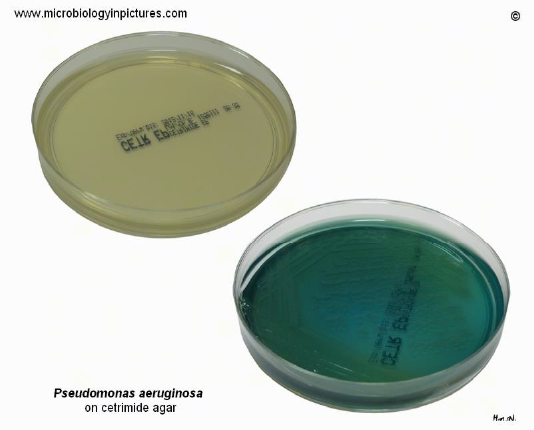
Figure 3: Cetrimide agar change its color from yellow to blue-green while showing growth of P. aeruginosa. Cultivation 24 hours at 37°C + 48 hours at room temperature (Banerjee et al., 2017).

Figure 4: Isolation of Mycobacterium bovis in stone brink media, yellowish and granular with a creamy appearance of M. bovis colonies (https://www.researchgate.net).
Mycoplasmas needs highly nutritive selective media for isolation as the media are enriched with beef heart infusion, inactivated horse serum, yeast extract, peptone, and other supplements (McVey et al., 2013). The agar was rendered selective by the addition of selective agents as antimicrobials such as thallium acetate or antibiotics (Miles and Nicholas, 1998). The samples were incubated at 37°C and 5% CO2 for 7–10 days, positive culture plates of Mycoplasma species revealed the growth of micro-colonies with a characteristic fried eggs appearance which can be detected by using light microscope (Quinn et al., 2013). The fried eggs appearance is due to the central dark zone of the colony inserting itself into the agar surrounded by a lighter peripheral zone of surface growth (Figure 5).

Figure 5: Microscopic image of Mycoplasma bovis colonies as fried eggs appearance (central dark zone surrounded by a lighter peripheral zone) (Ismael et al. 2018).
For the bacteriological culture of Brucella, selective agar media is required with the following composition; Brucella selective agar base, sterile sheep serum and antibiotics (polymyxin B sulphate, bacitracin, nystatin, cycloheximide, nalidixic acid, and vancomycin). The serum was boiled at 55°C for 30 minutes, filtered with a 0.2 μl sterile filter, and then mixed aseptically with Brucella agar base. The bacteria were isolated from milk samples using Brucella selective agar. Bovine milk samples were separately streaked onto Brucella selective agar media and inoculated culture media were placed in a CO2 incubator supplied with 5% CO2 at 37°C for 3–7 days (Islam et al., 2018). The manipulation of clinical specimens for the isolation of Brucella spp. was performed in a biosafety class II A2 cabinet. Resultant colonies were subcultured and identified by cultural characteristics and Gram staining (Figure 6).
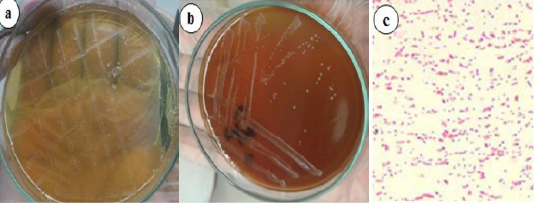
Figure 6: Brucella growth on selective agar media, blood agar media (a) Small, translucent, dewdrop-like round, and convex growth with smooth margins on Brucella selective agar media. (b) Whitish-grey, shiny, circular, convex, and nonhemolytic colonies of bacteria on blood agar media. (c) Gram-negative paired coccobacilli under a light microscope (400x) (Islam et al., 2018).
Molecular diagnosis of bacteria in bovine milk
Polymerase chain reaction (PCR)
Polymerase chain reaction is a revolutionary highly sensitive technique. Compared with conventional bacterial culture methods, the advantages of PCR technique is that it can operate directly using milk samples, and also it is less time consuming (Amin et al., 2011). Moreover, PCR is based on DNA and it doesn’t require the presence of live organisms. On the other hand, PCR has limitations as its profound sensitivity can yield false positive results (Yamagishi et al., 2007). A variety of PCR based tools have been developed for identification of infectious bacteria in bovine milk samples. The amplification of 16S and 16S-23S spacer rRNA genes is used for identification of Mycoplasma bovis, Staphylococcus spp., Escherichia coli and Streptococcus spp.(Hotzel et al., 1996; Amin et al., 2011). The amplification of 16S-23S rRNA intergenic spacer region (ISR) by PCR test is used for detection of Streptococcus agalactiae (Jiusheng et al., 2008) and Staphylococcus aureus genotype B (GTB) (Syring et al., 2012). Also, PCR test can be used for detection of Coxiella burnetii by amplification of transposon-like sequence gene which is found exclusively in C. burnetii (Willems et al., 1994). PCR used for specification of the antibiotic resistance genes of S. aureus, S. agalactiae and coagulase-negative staphylococci (CNS) which isolated from bovine milk (Ding et al., 2016; Osman et al., 2016). Also, PCR used for detection of virulence factors contribute to pathogenicity of E. coli, S. agalactiae, S. aureus, K. pneumonia, P. aeruginosa which are isolated from mastitic cow milk (Ding et al., 2016; Neamah, 2017). PCR techniques have become tool for diagnosis of Mycobacterium bovis in bovine milk. These techniques are less time consuming particularly in case of isolation of Mycobacterium bovis which needs more than six weeks of incubation (Bermudez et al., 2010). Moreover, PCR is used in epidemiological studies of bovine tuberculosis transmission. PCR technique is used to amplify a fragment of a 500-base pair of the RvD1-Rv2031c region which is confined to Mycobacterium bovis strains (Rahman et al., 2015) and gives a correspondence of 100% with the microbiological identification. Polymerase Chain Reaction is considered as a gold standard test for diagnosis of Brucellosis. Nested PCR is superior to conventional PCR due to its’ better sensitivity and specificity and it is capable of identifying a larger number of samples (Alwathnani et al., 2012).
Multiplex PCR
Multiplex PCR is a revolutionary molecular biology technique that is capable of detection of different types of organisms in one reaction by amplification of several different DNA targets in one set. On the other hand, it has limitations viz., low sensitivity which is attributed to the competition between different sets of primers (Amin et al., 2011). Also, multiplex PCR is utilized for detection of various infectious bacteria in bovine milk samples (Ammar et al., 2016). Multiplex polymerase chain reaction was used for direct detection of Mycoplasma bovis in cow milk samples after centrifugation and treatment of milk samples with mycoplasmal lysis buffer. The sensitivity of this PCR method ranged from 110 to 1,400 colony forming units (CFU). A main advantage of this method is the ability to detect Mycoplasma in milk samples from cows at an early stage of mycoplasmal mastitis where a small amount of mycoplasma could be detected in milk without culture. Moreover, Multiplex polymerase chain reaction is faster than conventional culture methods (Hirose et al., 2001). In a study, multiplex PCR was used for monitoring the presence of zoonotic tuberculosis in 181 raw bovine milk specimens from small farms (Bhanu Rekha et al., 2015); they were capable of differentiation between Mycobacterium tuberculosis and Mycobacterium bovis. They identified three positive samples for Mycobacterium tuberculosis and one for Mycobacterium bovis.
Real time PCR
Real time PCR is a highly specific quantitative technique that can both quantify and detect the bacterial pathogens in milk samples simultaneously. The most brilliant effect of this assay is its ability to overcome the concentration limit of conventional PCRs, since it can efficiently detect any nucleic acid even in very minute concentration. Moreover, it can determine the stage and type of infection. Therefore, it is more sensitive, specific, and rapid than conventional PCRs, and despite any drawbacks recorded, it is the technique of choice for laboratory diagnosis and molecular characterization (Zakary et al., 2011). Real-time PCR were used for bacterial identification in bovine milk samples from mastitic cows and also to detect the bacterial species that may fail to grow in conventional bacteria culture. These bacteria are Staphylococci spp., Streptococcus uberis, Streptococcus dysgalactiae, Corynebacterium bovis, Staphylococcus aureus, Escherichia coli, Enterococcus spp., and Arcanobacterium pyogenes (Taponen et al., 2009). Comparison between the real-time PCR assay and conventional culture technique for detection of bovine mastitis bacteria revealed that the real time PCR kit was superior to conventional culture technique in terms of speed, objective and automated interpretation of results, and high sensitivity in bacterial detection (Koskinen et al., 2010). Consequently, real time PCR is a promising innovative technique which can be used in parallel with conventional methods in diagnosis of mastitis. Real-time PCR assay was used to provide a rapid and sensitive method for the specific detection of Staphylococcus aureus in bovine milk (Zakary et al., 2011). Also, real-time PCR assay targeting multicopy insertion sequence (IS1111) was reported to measure amounts of C. burnetii shed in bulk milk samples (Kim et al., 2005). Eleven genes including interleukin 6, interleukin 8, cluster of differentiation 14, interleukin 1 beta, cluster of differentiation 14, acute-phase proteins, toll-like receptor 2, toll-like receptor 4, lipopolysaccharide binding protein, tumor necrosis factor and interferon showed high expression from two to four times during mastitis. These findings suggest that cows suffered from mastitis produced cell-mediated immune response (Zheng et al., 2006).
Pulsed field gel electrophoresis
Pulsed field gel electrophoresis (PFGE) was applied since 1980 and has been broadly utilized for strain typing and identification. During PFGE technique, the chromosomal DNA is first digested across a small number of sites along the chromosome using a restriction enzyme, producing large DNA fragments, then these fragments are separated in an agarose gel producing band patterns specific to different strains (Kaufmann, 1998). Pulsed-field gel electrophoresis (PFGE) performed on chromosomal digests of all Mycoplasma bovis isolates obtained from milk, mammary gland parenchyma, and supra-mammary lymph nodes in dairy cattle. The results confirmed that Mycoplasma strains isolated from the milk of dairy cattle with Mycoplasma mastitis frequently have PFGE patterns identical to those for strains isolated from other body sites, suggesting that there is an internal transmission of Mycoplasma organisms (Biddle et al., 2005). Using of PFGE in isolation and typing of subspecies or strain levels of mastitogenic pathogens, where coagulase (Coa) and protein A (Spa) genes of S. aureus strains were molecular typed. Also, the genetic heterogeneity between 19 pulso types of the strain was recorded. Thus, PFGE was found to be the best discriminatory assay for distinguishing strains (Ciftci et al., 2009).
Amplified fragment length polymorphism (AFLP)
Amplified fragment length polymorphism is a PCR-based technique also used for DNA fingerprinting, which involves digestion of the DNA with restriction enzymes, ligation of adaptors, and selective amplification of a subset of restriction fragments followed by gel analysis of these fragments (Vos et al., 1995). This technique identifies the sources of infection and transmission of the pathogen and studied the genetic variations of Mycoplasma bovis strains which were isolated from mastitic cattle in Denmark over a 17-year period from different geographic regions by utilizing the amplified fragment length polymorphism (AFLP) fingerprinting technique (Kusiluka et al., 2000). The insertion sequence IS6110 is considered specific to the members of the Mycobacterium tuberculosis complex and the difference in location and number of copies of the insertion sequence is a source of polymorphism between isolates. This polymorphism is detected by restriction fragment length polymorphism (Haddad et al., 2004).
Multi-locus sequence typing (MLST)
MLST tool is utilized in multiple loci sampling for discrimination between different strains of bacteria. It operates on a very simple principle as its’ procedure includes PCR amplification followed by DNA sequencing (Rabello et al., 2007). MLST utilized for the characterization of Mycoplasma bovis isolates from different geographical regions. The samples were collected from both healthy and infected cattle. They used 7 housekeeping genes namely; dnaA, metS, recA, tufA, atpA, rpoD, and tkt. MLST showed not only high discriminatory power but also revealed the presence of genetical differences among isolates mostly associated with geographical origins (Rosales et al., 2015).
Random amplification of polymorphic DNA (RAPD)
RAPD technique is based on utilization of a set of universal short primers in the interest of genome amplification by PCR, consequently, the resulting fragments are subjected to gel electrophoresis which leads to the formation of a semi specific profile or fingerprints. The difference of fingerprints among different isolates is attributed to minimal variation in the primer annealing site. This technique is advantageous in the terms of high speed and its capability of differentiating between a wide range of microbial species and their isolates (Kumari and Thakur, 2014). Streptococcus agalactiae genotypes isolated from bovine clinical mastitis by utilizing random amplified polymorphic DNA (RAPD) analysis. The Results showed a great genotypic diversity of Strep. agalactiae (45 RAPD-types) and three clusters (Ia, Ib and II) resulted from genetic similarity among genotypes. After clustering, high genetic similarity was recognized within and between herds (Tomazi et al., 2018).
Variable number tandem repeat (VNTR)
Tandem repeats are allelic variations that occur as repetitions of sequences of nucleotides found in intergenic regions of the genome. These repeated sequences are known in eukaryotic cells. The hypervariability of these repetitions in human and animal cells generates the variable number tandem repeat (Pitt and Barer, 2012).
In the diagnosis of mycobacterial isolates, the polymorphism may be analyzed by repetitions of sequences of nucleotides, or even variation in the number of repeating units (Rocha et al., 2013). The identification of 24 VNTR loci of M. tuberculosis genome received the name Mycobacterial Interspersed Repetitive Units (MIRU), and it has been used as a molecular marker for diagnosis of mycobacterial infections. Identification and classification by MIRU-VNTR method has proven to be fast, sensitive and affordable particularly for isolated mycobacteria (Supply et al., 1997). Twenty-five tandem repeats identified from 24 Streptococcus uberis strains which were isolated from the milk of mastitic cows (Gilbert et al., 2006), the study reported that advantages of this technique are easy to perform, inexpensive and can be applied in routine examination; help the investigations of the epidemiology of S. uberis mastitis in dairy cows.
Whole genome sequencing
Whole genome sequencing (WGS) is becoming a widely used assay for investigating bacterial genome sequences as high throughout sequencing becomes faster, cheaper, and more readily available. WGS involves sequencing the entire genome of selected isolates which can then be used for clinical diagnostics, disease outbreak investigation and controlling antimicrobial resistance (Koser et al., 2012). Whole genome sequence is completed for the major Gram-positive bacteria (S. aureus). The availability of completely sequenced genomes of S. auerus allows elucidation of amino acid sequences and identification of dominant epitopes of newly discovered mastitogenic pathogens virulence factors to be used in the design of vaccines for better control of mastitis (Baba et al., 2008). The complete genome sequences are available for 5 Mycoplasma bovis isolates (Sun et al., 2018). Comparing the genetic diversity between 82 Mycoplasma bovis isolates which were collected from various geographical locations by using WGS. They suggested the existence of a single strain. Minimal variation was reported between isolates from different geographical locations. In addition, minimal variation was also observed between isolates from clinically infected and subclinical carrier animals. They suggested that the disease outcome may be determined mostly by the host and environmental factors, while the pathogen factors playing a less significant role (Parker et al., 2016).
Immunological diagnosis of bacteria in bovine milk
Enzyme linked immunosorbent assays (ELISA)
Enzyme linked immunosorbent assays (ELISA) has been used to detect antibodies in bovine milk samples against many infectious bacteria such as Staphylococcus aureus (Fox and Adams, 2000), Listeria monocytogenes (Kalorey et al., 2007) and Leptospira hardjo (Rypuła et al., 2014). ELISA test is also used for measuring the levels of acute phase proteins as hepatoglobulin in milk which is considered as inflammatory marker of the milk during subclinical mastitis (Eckersall, 2007). The accuracy of a commercial milk amyloid A-ELISA (MAA-ELISA), bacteriological culture was estimated and SCC for diagnosis of subclinical mastitis (Jaeger et al., 2017). Also, the levels of MAA in normal milk and in subclinical mastitis cow’s milk was measured and it was recommended that the application of MAA concentration in milk to monitor the udder health in dairy cattle farms. ELISA assay used for evaluating the levels of three different acute phase proteins (APP) in bovine milk namely: Haptoglobin, C-reactive protein and mammary associated serum amyloid A3. The etiological factor of mastitis greatly influences the pattern of APP in milk, for instance, E. coli, S. uberis and S. dysgalactiae mastitis showed higher values of APP in milk as compared to other pathogens (Thomasa et al., 2018).
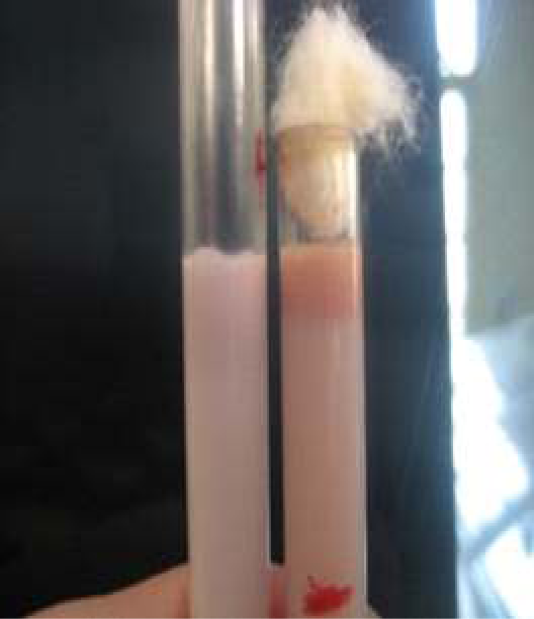
Figure 7: Milk ring test for detection of anti-Brucella IgM and IgA bound to milk fat globules. (Bhanu Rekha et al., 2015).
Milk ring test (MRT)
The milk ring test is capable of detecting antibodies to Brucella infection in milk. The main advantage of this test is that being inexpensive, simple and convenient, and does not need any special equipment to perform. It detects anti-Brucella IgM and IgA bound to milk fat globules. On the other hand, it has some limitations including false positive results which arise when milk contains colostrum or during the end of the lactation period. In addition, false reactions may also arise in cases of hormonal disturbance in cows or in cases of mastitis. The test can’t detect low concentrations of IgM and IgA attributed to its reduced sensitivity (Cadmusa et al., 2008) (Figure 7).
Conclusion and Recommendation
Raw bovine milk is considered as a vehicle for many infectious bacterial diseases such as Bovine Tuberculosis, Brucellosis, Leptospirosis, Q. fever and Mycoplasma infection. In addition to, Infectious bacteria associated with mastitis including Staphylococcus aureus, coagulase-negative staphylococci, Streptococcus dysgalactiae., Streptococcus agalactiae, Corynebacteriun pyogenes, Streptococcus uberis, Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumonia, Streptococcus pyogenes, Clostridium perfringens and Salmonella species were also included. Diagnosis of bacterial infectious diseases in bovine milk could be achieved by using conventional field tests, phenotypic diagnosis, recent developed molecular techniques, as well as serological diagnosis techniques. There are many available conventional and modern techniques used for the diagnosis of infectious bacteria in bovine milk. Conventional field tests such as somatic cell count, California mastitis test, electrical conductivity and pH tests have many advantages as they are convenient, non-expensive and can be used to monitor the udder health of the cattle. Microbial culture is a traditional technique which involves bacterial isolation, identification, Gram staining, microscopical examination, cultivation on enriched and selective media and studying biochemical characters. Microbial culture is fundamental diagnostic test for isolation of live infectious bacteria in milk. Modern molecular diagnostic techniques are more valuable and offer more sensitive, fast and highly confident results such as Polymerase chain reaction (PCR), Multiplex PCR, Real Time PCR and Whole genome sequencing. Genotyping diagnosis is widely used for genetic characterization of bacterial isolates such as pulsed field gel electrophoresis (PFGE), Amplified fragment length polymorphism (AFLP), multilocus sequence typing (MLST), Random amplification of polymorphic DNA (RAPD) and Variable number tandem repeat (VNTR). Another available diagnostic technique includes immunological diagnosis using Enzyme linked immunosorbent assays (ELISA) to detect not only antibody in bovine milk samples against many infectious bacteria, but also it is used to evaluate the levels of acute phase proteins such as hapatoglobin, c-reactive protein and mammary associated serum amyloid A3.
Author’s contribution
The Authors worked cooperatively while collection of information related to this review article.
Conflict of interest
The authors have declared no conflict of interest.
Animal welfare statement
The authors confirm that the ethical policies of the journal, as noted on the journal’s author guidelines page, have been adhered to No ethical approval was required as this is a review article with no original research data.
References





