Journal of Animal Health and Production
Research Article
P and G Genotyping of Bovine Rotavirus Detected from Fecal Samples of Calves in Abidjan District, Ivory Coast (2015-2017)
Jacques Danho Monney1*, Valery Edgard Adjogoua2, Yahaya Karamoko1, Marius N’takpé Adagba2, Agbaya Akran2
1Nature Sciences Training and Research Unit, Animal Biology and Cytology Laboratory, Nangui Abrogoua University. 02 BP 801 Abidjan 02, Ivory Coast; 2Virology departement of Institut Pasteur, 01 BP490 Abidjan 01, Ivory Coast.
Abstract | Bovine Rotavirus (BRV) is the major cause of gastroenteritis and diarrhea in newborn calves. Every year, Rotaviruses cause enormous economic losses. In current investigation, a total of 145 fecal samples were collected from both diarrheic (n=135) and non-diarrheic (n=10) neonatal calves. Samples were collected from diarrheal calves up to the age of three months, from bovine farms located in Abidjan district during 2015 to 2017. All the samples were screened by using conventional One step reverse transcription polymerase chain reaction (RT-PCR) for VP7 and VP4 genes. The results exhibited that 34.8% (47/135) samples were positive for both VP7 and VP4 genes. All non-diarrhoeic samples were negative for BRV. G10 was the most prevalent genotype, accounting for 61.7% (29/47). The detected P types were P[11] and P[5], accounting for 36.1% (17/47) and 23.4% (11/47) respectively. The most common genotype circulating among bovine population of Abidjan district is G10P[11] for 34% (16/47) and followed by G10P[5] for 8.5% (4/47). The results of this study suggest that these two types can be used as dominant strains for the formulation of an appropriate vaccine against bovine Rotavirus in the Abidjan district, Ivory Coast.
Keywords | Bovine, Rotavirus, RT-PCR, Genotype, Abidjan.
Received | June 20, 2019; Accepted | September 02, 2019; Published | September 15, 2019
*Correspondence | Jacques Danho Monney, Nature Sciences Training and Research Unit, Animal Biology and Cytology Laboratory, Nangui Abrogoua University. 02 BP 801 Abidjan 02, Ivory Coast; Email: monneyjacques@yahoo.fr
Citation | Monney JD, Adjogoua VE, Karamoko Y, Adagba MN, Akran A (2019). P and G genotyping of bovine rotavirus detected from fecal samples of calves in Abidjan district, Ivory Coast (2015-2017). J. Anim. Health Prod. 7(3): 106-112.
DOI | http://dx.doi.org/10.17582/journal.jahp/2019/7.3.106.112
ISSN | 2308-2801
Copyright © 2019 Monney et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Rotaviruses are important etiological agents of acute viral gastroenteritis in young one of many animals’ species and human (Trojnar et al., 2013; Kun et al., 2016). Rotavirus infections cause significant economic losses in neonates of many domestic animals. Neonatal calf mortality in the first month of age is accounted to be 80 to 85% of the total mortality and is particularly high in the third week of life (Uetake, 2012; Abebaw et al., 2018).
Rotaviruses are classified in the genus Rotavirus in Reoviridaea family. They are characterized by segmented genome including 11 segments of double stranded RNA contained within a triple layered protein shell composed of a core, inner capsid and outer capsid. Three structural proteins, VP1, VP2 and VP3, form the core of the Rotavirus particle (Gazal et al., 2011; Matthijnssens et al., 2011). Group A Rotaviruses are classified into G and P serotypes on the basis of outer capsids glycoprotein VP7 and VP4. Currently, 27 G-genotypes (from G1 to G27) and 35 P-genotypes (from P[1] to P[35]) have been identified in human and various species of animals in the world (Kapikian et al., 2001; Matthijnssens et al., 2012). Among these 27 G-genotypes and 35 P-genotypes, 11 G types (G1-G8, G10, G11 and G15) and 6 P-Types (P[1], P[5], P[11], P[14], P[17] and P[21]) have been reported in bovine Rotavirus (RV) infection with G6, G8, G10, P[1], P[5] and P[11] predominating (Adah et al., 2003; Garaicoechea et al., 2006; Beg et al., 2010). Some studies on calves in Nigeria and Tunisia have reported the presence of G8 genotype. The work undertaken by the African Rotavirus Network
Table 1: Primer sequences used for VP4 genotyping
| Primer | Sequence (5’-3’) | Position | Size | References |
| P-typing : full-length VP4 | ||||
| VP4-F | TAT GCT CCA GTN AAT TGG | 132-149 |
663bp |
(Gentsch et al., 1992) |
| VP4-R | ATT GCA TTT CTT TCC ATA ATG | 775-795 | ||
| partial length VP4 | ||||
| pNCDV-P1 | CGA ACG CGG GGG TGG TAG TTG | 269-289 | 530 |
(Gouvea et al., 1994b) |
| pUK-P5-P5 | GCC AGG TGT CGC ATC AGA G | 336-354 | 463bp | |
| 3T-1-P6 | TGT TGA TTA GTT GGA TTC AA | 259-278 | 331bp |
(Gentsch et al., 1992) |
| 1T-1D-P8 | TCT ACT TGG ATA ACG TGC | 339-356 | 345bp | |
| pB223-P11 | GGA ACG TAT TCT AAT CCG GTG | 574-594 | 222bp | (Gouvea et al., 1994b) |
Table 2: Primer sequences used for VP7 genotyping
| Primer | Sequence (5’-3’) | Position | Size | References |
| G-typing : full-length VP7 | ||||
| VP7-F | ATG TAT GGT ATT GAA TAT ACC AC | 51-71 |
881bp |
(Iturriza-Gomara et al., 2004) |
| VP7-R | AAC TTG CCA CCA TTT TTT CC | 932-914 | (Iturriza-Gomara et al., 2004) | |
|
partial length VP7 |
||||
| FT5-G5 | CAT GTA CTC GTT GTT ACG TC | 779-760 | 729bp |
(Gouvea et al., 1994a) |
| DT6-G6 | CTA GTT CCT GTG TAG AAT C | 499-481 | 449bp | |
| HT8-G8 | CGG TTC CGG ATT AGA CAC | 273-256 | 223bp | |
| ET10-G10 | TTC AGC CGT TGC GAC TTC | 714-697 | 664bp | |
| BT11-G11 | GTC ATC AGC AAT CTG AGT TGC | 336-316 | 286bp | |
(ARN) consortium has determined G8 and G10 in South Africa (Khuzwayo et al., 2012; Fodha et al., 2005; Esona et al., 2010). In Côte d’Ivoire, one study was conducted in 2004, and G10 genotype was detected in diarrheal calves (Akran et al., 2010).
There is no information on G and P diversity of bovine Rotavirus circulating locally in Abidjan district. Indeed, reduction in calf mortality rate and the economic losses of livestock in less industrialized countries is primarily based on effective vaccination. The aim of the present study was to identify genotypes circulating among bovine population of Abidjan district by using polymerase chain reaction (PCR) based typing assays.
MATERIAL AND METHODS
Study Area
The study area is located in Abidjan district (south of Ivory Coast). Breeding system encountered in Ivory Coast is the extensive system. There are no specified exploitation type, we generally meet a mixed production (milk and meat). Four seasons are observed in south of Ivory Coast: two rainy seasons and two dry seasons.
Sampling
A total of 145 (135 diarrheic and 10 non-diarrheic) faecal samples were collected from bovine calves with age group up to 3 months. Samples were collected from cattle farms of different places of Abidjan district, Ivory Coast from June 2015 to May 2017. Stools were collected directly from the rectum using gloves in sterile flasks. The samples were transported in a frost containing cold accumulators and stored at -20 °C until further processing.
Viral Rna Extraction
For VP4 and VP7 genes amplification, the viral RNA was extracted from collected faecal samples using QIAGEN Kit (RNA Qiamp® Viral, cat No 52906, Germany) following the manufacturer’s protocol. Extracted RNA was stored at -20 °C until further use.
Primers
Type specific primers reported for most common genotypes were selected based on available literatures. Sequences of primers, their positions in genomic segment are presented in Table 1 and 2.
Rotavirus Detection
RT-PCR was performed to detect Rotavirus gene in the fecal samples. Reaction was carried out by conventional method using One Step RT-PCR Kit (QIAGEN®) with pairs of forward and reverse amplification primers (VP7-F/ VP7-R; VP4-F/ VP4-R) to amplify respectively full-length of VP7 and VP4 gene (Iturriza-Gomara et al., 2004; Simmonds et al., 2008).
Vp7 Genotyping
G-typing was performed using a pool of internal primers (Gouvea et al.,1994b) specific to G genotypes G5, G6, G8, G10 and G11 bovine Rotavirus genotypes in combination with a appropriate forward consensus primer (VP7-F).
The reaction was performed on a gradient thermocycler under the following conditions: reverse transcription at 50 oC for 30 minutes, followed by PCR with initial activation at 95 oC for 15 minutes, followed by 35 cycles of 94 oC for 1 minute, annealing at 48 oC for 45 seconds and extension at 72 oC for 1 minute, and a final extension of 72 oC for 10 minutes.
Vp4 Genotyping
P-typing was performed using a pool of internal primers specific to P genotypes P[1],P[5],P[6], P[8] and P[11] bovine Rotavirus genotypes in combination with the appropriate forward consensus primer VP4-F (Gentsch et al.,1993; Gouvea et al., 1994a).
The reaction was performed on a gradient thermocycler under the same previous conditions.
Analysis of Pcr Products
The amplified products were analyzed by electrophoresis on 1% agarose gel containing ethidium bromide for 40 min at 100 V. The amplicons were visualized under UV light using a UV transilluminator. Samples identified as positive were analyzed and photographed by a gel documentation system.
RESULTS AND DISCUSSION
All samples from non-diarrhoeic calves were found negative for G and P. Full length amplification of both VP4 and VP7 gene of RV was obtained with bands sizes of 663 bp and 881 bp respectively in 47/135 of all diarrhoeic samples (Figure 1 and 2). Overall prevalence was found to be 34.8% in diarrheic calves. This is in agreement with many epidemiological studies in herds around the world that showed that Rotavirus was present in 30-40% of fecal samples of calves with diarrhea (Fukai et al., 2002; Badaracco et al., 2012; Dhama et al., 2009).
G10 was detected in 61.7% (29/47) of positive samples with a bande sizes of 661 bp (Figure 3). These results con
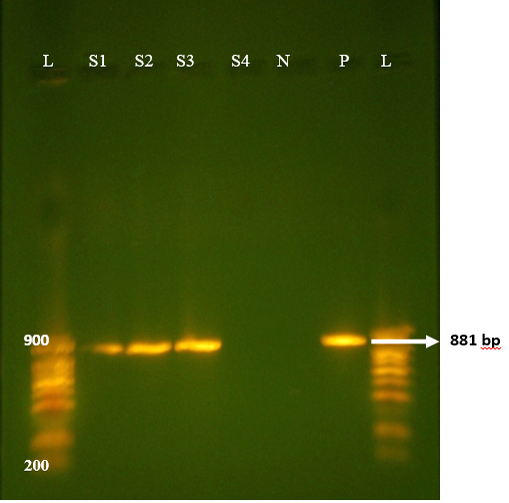
Figure 1: Agrose-gel electrophoresis of RT-PCR full length of VP7 (881bp) region of bovine Rotavirus. L: 100bp plus ladder, S1-S3: Positive faecal samples, S4: Negative faecal sample, N: Negative Control. P: Positive Control (Bovine RV).
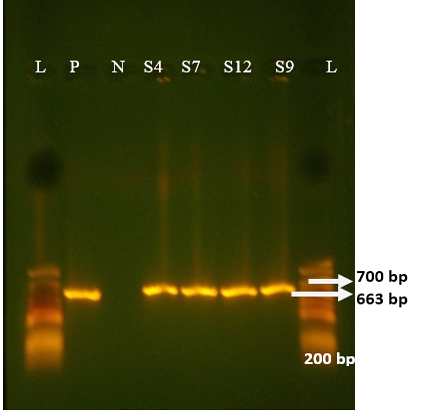
Figure 2: Agrose-gel electrophoresis of RT-PCR full-length of VP4 (663 bp) region of bovine Rotavirus. L: 100 bp plus ladder, S4, S7, S12, S9: Positive faecal samples, N: Negative Control. P: Positive Control (Bovine RV)
firmed the previous results obtained by Akran et al. (2010). G10 was the most predominant G type in this study (Table 3). Results found in our study is in agreement with reports in México, Iran and India (Rodriguez-Limas et al., 2009; Singh and Jhala, 2011; Prasad et al., 2015; Rajendran and Kang 2014; Nazoktabar et al., 2017); whereas in most countries such as Moroco, Hungary, Tunisia, Japan, Brazil and Europe, G6 was the most common genotype (Imane et al., 2016; Hajnalka et al., 2013, Fodha et al., 2005; Okada and Matsumoto 2002; Thabata et al. 2010; Sofie et al., 2012). G8 and G6 were not detected.
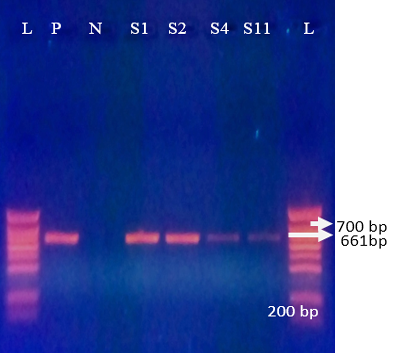
Figure 3: Agrose-gel electrophoresis of RT-PCR product of G10 (661 bp) genotype of bovine Rotavirus. L: 100 bp plus ladder, S1-S2, S4, S11: Positive faecal samples, N: Negative Control. P: Positive Control (Bovine RV)
Table 3: G and P genotypes in bovine stool samples from Abidjan district.
| Genotypes | +ve samples/ Number of samples (Percentage) |
| P/ G | 47/135 (34.8%) |
| G10 | 29/47 (61.7%) |
| P[11] | 17/47 (36.1%) |
| P[5] | 11/47(23.4%) |
| G10 P[11] | 16/47 (34%) |
| G10 P[5] | 8/47 (17%) |
Regarding P-genotypes, the most predominant P type was P[11] which was detected in 36.1% (17/47) with a band size of 222 bp (Figure 4) , followed by P[5] in 23.4% (11/47) with bands sizes of 463 bp (Figure 5). P[1], P[6] and P[8] were not detected. The present results are in accordance with those reported in many countries like Tunisia, Italy, france, Argentine, Turkey, similar to Iran, P[11] has been more frequently detected (Hassine-Zaafrane et al., 2014; Sofie et al., 2012; Badaracco et al., 2012; Alkan et al., 2010; Nazoktabar et al., 2017). These results are in contrast with others studies elsewhere in the world. In these countries like Ireland (Reidy et al., 2006; Cashman et al., 2010), Brazil (Alfieri et al., 2004), and India (Gantasala et al., 2014), P[5] is more common P-type compared to P[11].
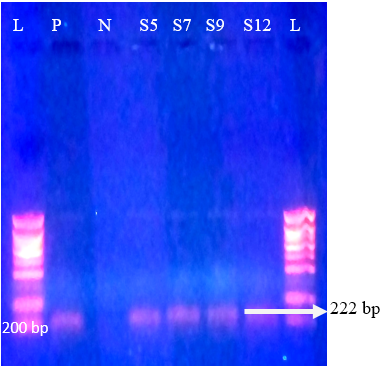
Figure 4: Agrose-gel electrophoresis of RT-PCR product of P[11] (222 bp) genotype of bovine Rotavirus. L: 100 bp plus ladder, S5, S7, S9, S12: Positive faecal samples, N: Negative Control. P: Positive Control (Bovine RV)

Figure 5: Agrose-gel electrophoresis of RT-PCR product of P[5] (463 bp) genotype of bovine Rotavirus. L: 100 bp plus ladder, S9, S22, S41, S53: Positive faecal sample. N:Negative Control. P: Positive Control (Bovine RV)
The combination G10P[11] was detected in 34% (16/47) followed by G10P[5] for 17% (8/47) of tested samples (Table 3). Similar G-P genotype combination was the most predominant combinaison and also recorded from different parts of the world: Tunisia (Hassine-Zaafrane et al., 2014), India (Ahmed et al., 2017; Prasad et al., 2015), France and Italy (Sofie et al., 2012), Japan (Abe et al., 2009), Iran (Nazoktabar et al., 2017). No other G-P combination has been found in our study. However according to the latest reports in many countries such as Ireland (Reidy et al., 2006; Cashman et al., 2010), Australia (Swiatek et al., 2010), Argentina (Badaracco et al., 2012) and South of Africa (Khuzwayo et al., 2012); G6P[5] was the most predominant combination. Otherwise, in Brazil, Thabata et al. (2010) and Fernanda et al. (2012) found G6P[11] the most dominant in 16.2%.
In India and Ivory Coast, authors have reported the presence of animal Rotavirus G10 in human gastroenteritis (Prasad et al., 2015; Akran et al., 2010). G10P[11] genotype was found to be present in bovine population and this may be recognized as an important contributor to the diversity of RV found in human population (Prasad et al., 2015; Nazoktabar et al., 2017). This involvement of G10 Rotavirus in human gastroenteritis is a serious threat of zoonotic transmission of this virus.
Using VP7F/VP7R and VP4F/VP4R primers designed respectively by Iturriza-Gomara et al. (2001) and Simmonds et al., (2008), 80% of samples that previously untypeable using con3/con2; sBeg/Beg; End9/EndA and BovCom3/ BovCom5 (Gouvea et al.,1990; Gentsch et al.,1992; Gault et al., 1999; Isegawa et al., 1993) primers could be typed. Primers designed by Iturriza-Gomara et al. (2001) and Simmonds et al. (2008) increased the sensitivity of genotyping,comparing with others. This result is in agreement with that of Nirwati et al. (2013) in Indonesia. These authors concluded that primers designed by Iturriza-Gomara et al. (2001) and Simmonds et al. (2008), were more sensitive than those described by others.
CONCLUSION
The present study reveals that G10P[11] is predominant type of RV circulating in bovine population in Abidjan district. G10 poses a serious threat to humans because of zoonotic nature of the virus. The widespread surveillance of animal and human population is needed to determine the diversity of Rotavirus circulating in Ivory coast before adopting any immunization programs.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the technical staff of Virology Departement of Institut Pasteur, Ivory Coast.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
Authors Contribution
All authors contributed equally.
REFERENCES





