Journal of Infection and Molecular Biology
Research Article
Diagnosis of Babesia Bovis Infection in Indigenous and Crossbred Cattlewith Comparison Between Conventional and Molecular Diagnostic Techniques
Shakeel Hussain1, Kamran Ashraf2, Naveed Anwar3, Muhammd Ameen Jamal4, Huma Naeem2, Nisar Ahmad2, Aziz-Ul-Rahman5*
1Department of Wildlife Conservation and Utilization, Northest Forestry University Harbin 150069, China; 2Department of Parasitology, University of Veterinary and Animal Sciences Lahore 54600, Pakistan; 3State Key Laboratory of Veterinary Biotechnology, Harbin VeterinaryResearch Institute, Chinese Academy of Agricultural Sciences, Harbin 150069, China; 4Department of Animal Breeding Genetics and Reproduction, Yunnan Agricultural University, Kunming 650201, Yunnan, PR China; 5Department of Microbiology, University of Veterinary and Animal Sciences Lahore 54600, Pakistan.
Abstract | A comparative study was conducted to evaluate the incidence of Babeisa bovis (B. bovis) infection in crossbred and indigenous cattle with relation to age and breedin Lahore, Pakistan using microscopic examination and PCR technique. A total of 60 blood samples of clinically ill cattle were obtained from different dairy farms. An overall prevalence of 8.3% by PCR and 6.67% by microscopy were recorded for B. bovis. The crossbred cattle showed significantly (p<0.05) high prevalence of 14.33% and 10% by using PCR and microscopy, respectively, compared to indigenous cattle with 3.33% using both diagnostic techniques. Among the different age group, animals with >2 year of age had a higher prevalence of 15.38% than 5.26% in 1-2 year of animals and at least 0.00% in <1 year of animals. It was concluded from this study that PCR found as an efficient technique rather than microscopy for diagnosis of babesiosis in carrier state infectionin those regions where haemoparasites are endemic in the cattle population.The findings of this work may aid in the prevention or control of the babesiosis.
Keywords | Prevalence, Babesia bovis, Indigenous cattle, Crossbred cattle, Microscopy, PCR.
Editor | Tahir Yaqub, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | February 01, 2017; Accepted | March 03, 2017; Published | March 25, 2017
*Correspondence | Aziz-ul-Rahman, Department of Microbiology, University of Veterinary and Animal Sciences Lahore 54600, Pakistan; Email: drazizangel@gmail.com
Citation | Hussain S, Ashraf K, Anwar N, Jamal MA, Naeem H, Ahmad N, Rahman AU (2017). Diagnosis of babesia bovis infection in indigenous and crossbred cattlewith comparison between conventional and molecular diagnostic techniques. J. Inf. Mol. Biol. 5(1): 1-6.
DOI | http://dx.doi.org/10.17582/journal.jimb/2017/5.1.1.6
ISSN (Online) | 2307-5465; ISSN (Print) | 2307-5716
Copyright © 2017 Hussain et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
In Pakistan livestock sector is a major contributor in the national (11.8%) and agriculture (56.3%) economy with Rs. 801.3 billion gross value of livestock. Pakistan is 4th largest milk producing country with the fastest growing dairy sector from last several years as compared to the other countries (Economical Survey of Pakistan, 2014-15). Livestock production system is negatively affected by diseases, thus setting off a cascading effect of reduced production, low income, and subsistent livelihood (Singh et al., 2004). Genuinely, a vast range of native animals, including cattle, goat, sheep, horse, pigs and dogs have been observed as victim of tick born infections (Bock et al., 2004). Ticks infestation results in spread of serious pathogens like Babesiosis which is the utmosthealth snags of the livestock sector in developing countries (Aktas et al., 2012). It is caused by hemo-protozoan parasites of the genus Babesia (Spickler and Roth, 2008) and transmitted by ticks of the Ixodidae family (OIE, 2008) and Boophilus species (Khan, 1991) as a main vector globally. Regard to species diversity, amongsix well-known Babesia spp. (Altay et al., 2008; Razmi et al., 2013), Babesia bovis (B. bovis) have the highest impact onthe livestock sector (Uilenberg, 2006) because of less sensitive to some babesiacidal drugs, thus making it more problematic to treat the infected animals.
As the 2nd most common hemo-parasite after trypanosomiasis in mammals (Hunfeld et al., 2008), the B. bovis,that parasitizedthe blood erythrocytes (RBCs) results in haemolyticanaemia associated with impaction of rumen, loss of appetite, and nervous sign e.g. incoordination, mania and teeth grinding.Subsequently, the death can also occur as consequences of the involvement of central nervous system (Bock et al., 2004). The increment of pyrexia during infection may cause abortion in pregnant animals. Moreover, bulls also show a reduction in fertility which lasts for about 6-8 weeks (Talkhan et al., 2010). Bos indicus breeds almost perpetually experience slighter clinical signs to primary B. bovis infections than Bos taurus breeds (Hunfeld et al., 2008). This phenomenon is believed to have an evolutionary relationship between Bos indicus cattle and babesiosis (Hunfeld et al., 2008).
After recovery from babesiosis, the animals generally become carriers (Chaudhry et al., 2010) and considered as a reservoirfor natural transmission of the disease. However, serology for detection of the carrier state in animals lacks specificity and sensitivity, especially for the diagnosis of infection status (Iseki et al., 2010). But, for better diagnosis, there are various molecular assayshave been developedto detect the carrier state of infection without samples (Dey and Singh, 2006). For molecular diagnostic of babesiosis, the application of PCR assaysis still incipient, but recently a number of studies have been revealed the significance of PCR assays to detect and differentiate the various species of Babesia at carrier stage with high sensitivity and specificity (Oliveira et al., 2005; Buling et al., 2007; Ravindran et al., 2008; Singh et al., 2010).
Moreover, numerous molecular techniques have been developed, but the PCR has become as a rapid sensitive assaywhich are capable of genomic detection (Bloch et al., 2013). The PCR assay is believed to be a powerful tool for the diagnosis of babesiosis (Altay et al., 2008). The cattle have been the companion of human being since ages, and shares the environment and belongings. But, Babesia infestation as a main constraintfor livestock productivity and infecting the human resources directly and indirectly. The present study was designed for the establishment of future control strategies using a specific, reliable and sensitive molecular tool for the detection of B. bovis infection. Two different diagnostic methods, microscopy and PCR used and compared to detect parasite prevalence. Furthermore, the findings of current study also provided a baseline factsab out the incidence of B. bovis and risk factors involved.
Materials and methods
Sample Collection
A total of 60 blood samples 60 clinically illcattle [Indigenous cattle (n=30) and crossbred cattle (n=30)] were collected with the reference of the cardinal signs (Pyrexia, Haemoglobinuria, Anorexia, Lethargy with malnutrition)from various dairy farms situated in Lahore region. The blood was collected aseptically via the jugular vein in anticoagulant (3.2%) coated tubes and stored at 4ocfor future analysis. In order to evaluate the risk factors (species, age, presence of ticks and herd size), the datawere obtained through questionnaires completed by investigators on sampling sites.
Microscopic Examination and Molecular Detection
For microscopy, using glass slide the blood smears of all samples were prepared and fixed with methanol for Giemsa staining as follows by (Ahmad et al., 2014) and observed microscopically. For DNA extraction, the blood samples were further stored at -20oC and whole-genomic DNA was isolated from blood sample using GeneAll DNA blood extraction kit as following the protocol. For molecular detection of B. bovis,the polymerase chain reaction (PCR) was carried out using the sequences of oligonucleotide primers specific for B. bovis as described by (Zulfiqar et al., 2012). The sequences of the primers are as follows
Forward primer:5ʹ-CTGTCGTACCGTTGGTTGAC-3’
Reverse primer: 5’-CGCACGGACGGAGACCGA-3’
The PCR products were checked for amplification by electrophoresis on a 1.5 % agarose gel and visualized under ultraviolet light. In order to check the specificity of the assay, genomic DNA of B. bovisisolated from the microscopically positive cases by standard protocols were also employed in the PCR to see the amplification, if any. The results of PCR assay were compared with that of Giemsa-stained blood smear examination.
Statistical Analysis:
The data was statistically analysed by applying Chi square test using Statistical Package for Social Services (SPSS) version 20.0.The P value was calculated, where 0.05 was considered significant at the 95% C.I (Confidence Interval) level.
RESULTS
The present study was conducted in Lahore to focus the prevalence and molecular detection of Babesia bovis in cattle population presenting at the dairy farms.Out of these 60 blood samples, 6.67% and 8.3% positivity have been observed for the existence of B. bovis by microscopy and PCR techniques (Table 1).
Table 1: Comparative prevalence rate of Babesia bovis among designated cattle population in Lahore
|
Diagnostic Technique |
No. of sample |
Positive No. |
Prevalence |
P-value |
|
Microscopy |
60 |
4 |
6.67% |
0.000* |
|
PCR |
60 |
5 |
8.3% |
*=Significant (p<0.05), PCR=Polymerase Chain Reaction
The microscopic blood smear examinationrevealed positive samples for B. bovis infected animals paired or single pear-shapedtrophozoites (pyriform), intra-erythrocytic parasite appearance (Figure 1). Regards to comparison in indigenous and crossbred cattle, microscopy showed 3.33% with 1 positive sample in indigenous cattle and 10% with 3 positive samples in crossbred cattle (Table 2). Among different age groups of animals, the animals of >2 age have been observed in most positive (15.38%) followed by 0.0% for1-2 year and <1-year age of animals (Table 3).
Table 2: Incidence of B. bovis in native and crossbred cattle
|
Breed |
No. of sample |
Microscopy |
PCR |
||
|
Positive |
P- value |
Positive |
P- value |
||
|
Indienous Cattle |
30 |
1(3.33%) |
0.002* |
1(3.33%) |
0.010* |
|
Crossbred Cattle |
30 |
3(10%) |
4(14.33%) |
||
*=Significant (p<0.05), PCR= Polymerase Chain Reaction
Table 3: Age-wise distribution of B. bovis infestation in cattle population of various dairy farms
|
Age Group |
No. of sample |
Microscopy |
PCR |
||
|
Positive |
P- value |
Positive |
P- value |
||
|
<1 year |
15 |
0(0.0%) |
0.061NS |
0(0.0%) |
0.193NS |
|
1-2 year |
19 |
0(0.0%) |
1(5.26%) |
||
|
>2 year |
26 |
4(15.38%) |
4(15.38%) |
||
NS=Nonsignificant (p>0.05), PCR=Polymerase Chain Reaction
Further, in order to evaluate the true status of B. bovis infection, all samples were analysed by PCR.Of the total 60 samples, 5 (8.3%) were found to be positive for B. bovis infection as revealed by the amplification of a 491-bp product (Figure 2). The results reported the high incidence of 14.33% in crossbred cattle as compared to indigenous cattle with 3.33% (Table 2).The animals with >2 year of age showed high positivity (15.38%) than the 5.26% in 1-2 year and 0.0% in <1-year-old animals (Table 3).
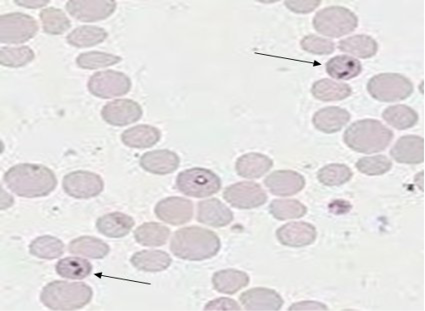
Figure 1: Giemsa stained microplate showing characteristics of babesia bovis infection. Thin arrow shows Babesia sp. inside the bovine erythrocytes

Figure 2: Representative gel showing the specificity of PCR for the efficient detection of B. bovisLane 1 & Lane 3: Negative samples, Lane 2 & Lane 4: positive sample for B. bovisat 491bp and Lane M, 1000bp DNA ladder marker
Discussion
In the present study, two different techniques (Microscopic examination and PCR) were used for detection of B. bovis in collecting samples, but previous studies reported PCR as an efficient technique rather than microscopy due to lack specificity and sensitivity, (Iseki et al., 2010). Because, in case of low parasitism or subclinical infestation of B. bovis remain undetectable microscopically (Ferreri et al., 2008; Schneider et al., 2011) but the PCR based technique was considered as more reliable for diagnosis of babesiosis in these animals and also for carrier state infection (Schneider et al., 2011) in numerous animal species of related countries, having similar climatic conditions, including Tunisia, United Arab Emirates and Iran (Zaeemi et al., 2011). As a higher out come in the present study are in agreement with previous study by (Altay et al., 2008) who reported PCR as more sensitive and specific tool than other conventional methods for the diagnosis of babesiosis. As similar findings, PCR has been noticed as more sensitive than microscopic blood smear examination in the diagnosis of the babesiosis in House hold dairy cattle (Shams et al., 2013). So, regards to comparison of sensitivity rate, a previous studyfrom India notified PCR (16.19%) at most reliable diagnostic tool to investigate of B. bovis as compared to blood smear techniques (1.8%) (Bhat et al., 2015). Another study was conducted for detection of babesiosis in cattle of Khyber Pakhtunkhwa Pakistan, high prevalence (27.5%) has been estimated by PCR as comparison with microscopy (9.83%) (Guido et al., 2002).
In the present study, indigenous and crossbred cattle were examined for existence of B. bovis cause of higher severity of infection has been observed in cattle than buffaloes (Mahmmod et al., 2008). In the present study, the overall prevalence was detected as 6.67% by microscopic examination and 8.3% by PCR, which are the similar findings of previous study, who reported the prevalence of 3% in the cattle population of District Kasur, Punjab, Pakistan (Durrani et al., 2008). Another study has shown the overall 18% positivity with 11% for B. bovis and 18% for B. bigemina (Chaudhary et al., 2010). But our findings are much lower than the previous documentation of babesiosis infection rates among cattle at 40% and 75% by microscopic examination and by PCR, respectively (Hazem et al., 2014), whereas, previous studies on the prevalence rate (7.10% and 9.3%) of B. bovis reported in Iran andErzurum region, respectively support the current study findings (Duzlu et al., 2015). In another previous study, which was conducted in Pakistan revealed the high prevalence of B. bovis as 19% in Charsada and 20% of Swabi districts (Ahmad et al., 2014). These findings are much higher than the present study’s findings and previous study of Egypt, where prevalence was 3.97% (Spickler et al., 2008). The influence of tick prevalence in distinguish areas is attributed to various factors including, geo-climatic conditions, awareness/education of the farmers, and farm management practices with lifestyle of different species of animals (OIE, 2008).
The findings of the current study about the influence of breed exposed that crossbred cattle are at most prone (14.33%) to babesiosis, as compared to indigenous cattle (3.33%) which are the similar results showed by a previous study (Bock et al., 2004) in native cattle. Conversely, (Velosamy et al., 2014) found crossbred cattle at more risk for prevalence of B. bovis than indigenous breed animal.Similar to these, In Pakistanthe studies of (Chaudry et al., 2010; Niazi et al., 2008) showed 11% and 7.2% prevalence, respectivelyamong crossbred cow as a comparison of indigenous cow.
The different risk factors such as age, sex and breed have been observed to influence on prevalence of haemoparasites (Kamani et al., 2010; Alim et al., 2011). To know the relation of babesiosis with the age of the cattle, the current work showed the higher prevalence rate (15.38%) of B.bovis in the cattle of higher age group. On the basis of age, a previous study from Ethiopia reported the highest prevalence among old animals (23.5%) followed by adult (15%) and young animals (13.2%) (Hamsho et al., 2015).The findings of the present study agree with (Velosamy et al., 2014) who founda similar trend and attributed theirfinding with higher infection rate of 8.33 % among the age groups of 2-7 years whereas in animals of below 2 years.
Conclusion
The present investigation proposed the PCR procedure as a valuable tool for the epidemiological analysis, evaluation and control planning about B. bovis infection incattle herds.Because, diagnosis of carrier animals has significant influence for preventing outbreaks in herd by transmission through vector ticks to healthy animals. Based on the current study’s findings, it is suggested that the tick eradicationprogram should be launched and farmer/owners of the animals should be trained to improve themanagement practices to reduce the pick burden on farms.This study focused only torecorded the prevalence of haemo-protozoa in fewdairy farms of Lahore, it is therefore suggested that further studies should be conducted in other dairy farms and to determine the efficacy of anti-protozoan drugs. A planned routine screening investigation should be conducted from different herds, especially from valuable cattle in regions where the haemo parasite species are concurrently found infecting bovines and need of immediate veterinary care to overcome the health problem of cattle relating to bovine B. bovis.
Acknowledgments
Authors are thankful to the Chairman, Department of Parasitology University of Veterinary and Animal Sciences Lahore, Pakistan for providing facilities to carry out this research work.
Author’s Contributions
All authors listed have made substantial, direct and intellectual contribution to this work and approved it for publication.
Conflict of interest
None of the authors of this study have a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of the paper.
References





