Journal of Infection and Molecular Biology
Research Article
Comparison of Two Deparaffinization Techniques and Three DNA Extraction Methods from Paraffin-Embedded Biopsies
Refka Jelassi1*2, Rym Ben Abdallah1, Hanen Chelbi1, Nissaf Ben Alaya3, Slim Haouet4, Aida Bouratbine1, Karim Aoun1
1Laboratory of Medical Parasitology, Biotechnology and Biomolecules, Pasteur Institute of Tunis, Tunisia; 2Faculty of Sciences of Bizerte, University of Carthage, Tunisia, 3National Observatory of New and Emerging Diseases, Tunisia; 4Pathology Department, La Rabta Hospital, Tunis, Tunisia.
Abstract | The formalin fixed paraffin-embedded tissues (FFPET) represent an important source of biological material. However, the quality of the extracted DNA from such biopsies is often poor probably because of the type of fixative, fixation duration, block age and persistence of paraffin residues. The objective of the current study was to compare two deparaffinization techniques in combination with three extraction methods in order to select the best one to be used in routine diagnosis. Two deparaffinization techniques; xylene and temperature were tested. They were combined with three commonly used extraction methods namely phenol-chloroform, the commercial kit QIAamp DNA Mini Kit (Qiagen) and the salting-out method. Thirteen samples of FFPET, among which three infected by Entamoeba histolytica, were used for this evaluation. The quantity and quality of obtained DNA were measured using NanoDrop. The IL6 human gene was also amplified for further validation. The combination temperature/commercial kit revealed the most efficient with good DNA yield and high purity (p≤0,001). It also provided the best success rate of PCR amplification. This method is simple and does not use toxic substances. Amplification products confirmed the presence of Entamoeba histolytica in 2 out of 3 amoebiasis biopsies tested. The combination temperature/commercial kit allows the DNA extraction with good quantities and high purity from FFPET. Such biological samples may represent a possible alternative in research and diagnosis.
Keywords | Deparaffinization, DNA extraction, PCR, IL6 gene, Entamoeba histolytica
Editor | Tahir Yaqub, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | June 29, 2016; Accepted | December 05, 2016; Published | December 13, 2016
*Correspondence | Refka Jelassi, Laboratory of Medical Parasitology, Biotechnology and Biomolecules, Pasteur Institute of Tunis, Tunisia; Email: refka.jelassi@gmail.com
Citation | Jelassi R, Ben Abdallah R, Chelbi H, Ben Alaya N, Haouet S, Bouratbine A, Aoun K (2017). Comparison of two deparaffinization techniques and three DNA extraction methods from paraffin-embedded biopsies. J. Inf. Mol. Biol. 4(3): 44-48.
DOI | http://dx.doi.org/10.14737/journal.jimb/2016/4.3.44.48
ISSN (Online) | 2307-5465; ISSN (Print) | 2307-5716
Copyright © 2017 Jelassi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
The Molecular biology techniques mainly those based on gene amplification are increasingly used in the field of research and in diagnosis (Mies, 1994). They often deal with blood, biological fluids and fresh or frozen tissues. The development of DNA extraction methods from formalin-fixed paraffin-embedded tissues (FFPET) enabled the use of such samples as possible alternative to fresh or frozen tissues (Salvador and Stephen, 1997). These samples provide many advantages namely a long preservation of tissue and an easy storage (Scicchitano et al., 2006; Rivero et al., 2006). Furthermore, Biopsies in paraffin blocks allow obtaining genetic material useful in many fields such as genomic and epidemiological studies. In addition, in certain clinical situations the paraffin embedded tissues are very useful for molecular diagnostic confirmation of certain diseases, identification of infectious agents such as viruses and parasites, genetic characterization of hereditary disease for a deceased subject or confirmation of malignancy, as well as cancer research for either retrospective and prospective studies (Mies, 1994). However, the quality and quantity of the DNA extracted from FFPET pose several problems because such DNA is often scare, degraded and can contain substances that inhibit the molecular procedure (Coura et al., 2006). In fact, several factors can negatively affect the DNA extraction from FFPET. We can cite the type of fixative, fixation duration, block age and persistence of paraffin residues in the DNA extract.
So that the importance of this genomic DNA source applicable in different fields explains the development of extraction and purification techniques which aims to optimize the quality and quantity of DNA extracted from this type of sample.
The objective of this study is to compare the results obtained by the combinations of two deparaffinization techniques with three extraction methods in order to choose the best one for routine activities.
Materials and Methods
Ten recent biopsies from different human tissues (uterus, prostate, endometrium, polyp, cutaneous biopsy) and three cerebral biopsies retained as infected by Entamoeba histolytica (anatomic-pathological diagnosis) have been used. They have been fixed in a solution of 10% formalin before being embedded in paraffin. For each sample, we used a section of ten microns to test the different extraction methods. These samples were provided by the Anatomic-pathology departments of the Pasteur Institute of Tunis and “la Rabta” hospital in the frame of the routine diagnosis activity.
Deparaffinization Methods
Two methods, based on xylene and temperature, were tested.
Chemical method: It was performed as describe by Goelz et al. (1985). Two xylene washes were done (1ml of xylene for 10 min at 56°C). The pellet was then washed in decreasing concentrations of alcohol (100%, 95%, 70%) to remove residual xylene. The samples were finally resuspended in water for molecular biology.
Thermal method: The samples were incubated with tissue lysis buffer (ATL) and proteinase K in a heating block at 65°C overnight. The tubes were centrifuged at high speed 14000 rpm for 5 min in refrigerated centrifuge (+4°C). The supernatant was then collected to be used.
DNA Extraction Methods
Three commonly used methods namely phenol-chloroform, QIAamp DNA Mini Kit (Qiagen) and the salting-out method were applied.
The phenol-chloroform method: The samples were incubated in digestion buffer (50 mM Tris-HCl, pH 8.0; 10 mM EDTA, pH 8.0; and 50 mM NaCl) containing 10% SDS and Proteinase K (10 mg/ml) at 65°C overnight. After digestion, saturated phenol was added and micro tubes were gently shaken by hand and centrifuged for 10 min at 4000 rpm. The upper phase was transferred to a new tube and mixed with phenol-chloroform-isoamylalcohol (1/24).
Commercial kit extraction: The extraction procedure was performed according to the manufacturer’s instructions (QIAamp DNA Mini Kit, Qiagen; Germany). After digestion with ATL buffer, the samples were incubated with AL buffer and Proteinase K (10mg/ml) at 70°C for 2-3 hours, and then mixed with 100% ethanol. The solution was transferred into a spin column, centrifuged for 1 min at 8000 rpm and washed with AW1 and AW2 buffers. DNA was eluted with AE buffer and stored at -20°C.
The salting-out method: The samples were incubated in digestion buffer (1 mM Tris–HCl, pH 8.0; 0.5 mM EDTA; and 0.5% Tween 20) and 20 mg/ml of proteinase K overnight at 56°C. The precipitation step was realized using 2M ammonium acetate and isopropanol. The pellet was washed with 70% ethanol and the extracted DNA was stored in TE buffer (1 mM Tris–HCl; 0.5 mM EDTA, pH 8.0) at -20°C (Funabashi et al., 2012).
Evaluation of the Extracted DNA
Amount and purity of DNA: The amount and purity of the extracted DNA were evaluated by spectrophotometer NanoDrop (Thermo Scientific NanoDrop 2000, USA). The DNA concentration (ng/µl) and the ratio of optical density OD260nm and OD280nm (R 260/280) were measured. DNA is considered pure when the ratio 260/280 is around 1.8.
PCR amplification of the IL6 human gene: The quality of the obtained DNA was also evaluated by PCR amplification using primers targeting a 105 bp fragment of IL6 human gene. IL6 is a human gene present in a single copy. In addition, the amplicon size used (105pb) is adequate for amplifying more copies view that we often cannot amplify DNA extracted from FFPET using long fragments. The primers used in our study were those described by Pereyra et al. (2012).
PCR reaction was carried out in a total volume of 25µl containing 1,5U Taq polymerase (AmpliTaq Gold; Applied Biosystems, USA), 1X PCR buffer, 25mM of MgCl2, 200µM of dNTPs, 20pmol of each primers (IL6 Forward5’-GCCTCAATGACGACCTAAGC-3’ and IL6 Reverse 5’-GGGGCTGATTGGAAACCTTA-3’) and 1µl of extracted DNA. The cycling conditions were as follows: initial denaturation at 94°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 62°C for 30 seconds, extension at 72°C for 30 seconds and a final extension step at 72°C for 5 minutes. The amplicons were electrophoresed using 2% agarose gel stained with red safe and visualized under ultraviolet light.
Table 1: Amounts and purities of DNAs extracted and time consuming of each method
|
Combinations |
DNA Amount (Mean±SD) (ng/µl) |
DNA Purity (260/280) |
Time consuming |
|
A: xylene/phenol-chloroform |
324,14 ± 46,22 |
1,68 ± 0,09 |
3-4days |
|
B: temperature/phenol-chloroform |
226,84 ± 36,05 |
1,75 ± 0,11 |
3-4days |
|
C: xylene/QIAamp DNA Mini Kit |
322,24 ± 79,56 |
1,90 ± 0,01 |
2days |
|
D: temperature/QIAamp DNA Mini Kit |
296,43 ± 48,08 |
1,85 ± 0,04 |
2days |
|
E: xylene/salting-out |
55,93 ± 14,41 |
1,23 ± 0,10 |
2days |
|
F: temperature/salting-out |
231,37 ± 48,96 |
1,61 ± 0,10 |
2days |
|
Friedman Test |
P=0,001 |
P<0,001 |
SD: Standard deviations
To verify the efficiency of the selected combination, we extracted DNA from three brain biopsies embedded in paraffin for which histological diagnosis revealed the presence of E. histolytica the agent of amoebiasis. The obtained extracts were amplified using the PCR method described by Gonin and Trudel (2003) which allows the differentiation of E. histolytica and E. dispar using ED1 and EH1 primers targeting a 135bp fragment of rDNA with the same reverse EHD2.
Statistical Analysis
Data obtained were submitted to variance analysis using SPSS version 20. The comparison between the different combinations was performed using the Friedman test. The methods were compared pairwise using ANOVA test. Differences were considered statistically significant when P-value ≤ 0.05.
Results
All methods provided a good amount of DNA excluding xylene/salting-out combination (Table 1). The highest amounts of DNA were obtained using phenol-chloroform and commercial kit methods either with xylene or temperature (Table 1).
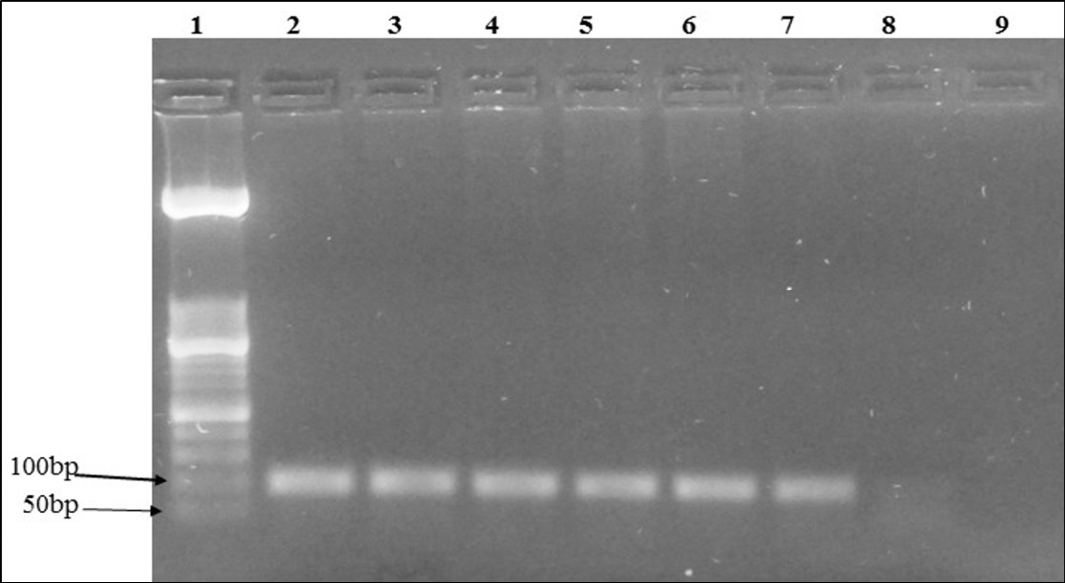
Figure 1: Agarose gel electrophoresis of IL6 human gene PCR of extracted DNA from the same sample using various combinations: Lane 1: molecular weight (50 bp); Lane 2: positive control; Lane 3 to 7: positive samples; Lane 8: negative sample; Lane 9: negative control
The DNA purity was statistically different between the six combinations (p<0,001) (Table 1). The commercial kit showed the highest purities (1,85 and 1,90) either with temperature and xylene respectively while the xylene/salting-out method provided statistically the least impurities when compared with other methods (p<0.05). The difference between the other methods, taken in pairs, was not significant.
Table 2: Results of PCR amplification of IL6 human gene
|
Combination |
A |
B |
C |
D |
E |
F |
|
Sample 1 |
+ |
+ |
+ |
+ |
- |
+ |
|
Sample 2 |
+ |
- |
+ |
+ |
- |
- |
|
Sample 3 |
+ |
+ |
+ |
+ |
+ |
+ |
|
Sample 4 |
+ |
- |
+ |
- |
+ |
+ |
|
Sample 5 |
- |
- |
- |
- |
- |
- |
|
Sample 6 |
- |
- |
- |
- |
- |
- |
|
Sample 7 |
- |
- |
- |
+ |
- |
- |
|
Sample 8 |
+ |
+ |
+ |
+ |
+ |
- |
|
Sample 9 |
- |
- |
- |
- |
- |
- |
|
Sample 10 |
- |
- |
- |
+ |
- |
- |
|
Percentage (%) |
50 |
30 |
50 |
60 |
30 |
30 |
+ : positive amplification; - : negative amplification ; A: xylene/phenol-chloroform; B: temperature/phenol-chloroform; C: xylene/QIAamp DNA Mini Kit; D: temperature/QIAamp DNA Mini Kit; E: xylene/salting-out; F: temperature/salting-out
The xylene/phenol-chloroform, xylene/kit and temperature/kit combinations showed the highest percentage of positive PCR amplification of the IL6 human gene (Table 2). As an example, the PCR amplicons of sample No. 1 obtained with the six combinations are shown in Figure 1. The phenol-chloroform method revealed the most time consuming (3 to 4 days) (Table 1). It was also laborious to perform.
According to the results, the combination of temperature/Qiagen kit is considered as the best in terms of providing DNA with good quantity and quality. It was selected and used to extract DNA from the three brain FFPET corresponding to confirmed cases of cerebral amoebiasis. PCR amplification showed that two out of the three tested samples revealed the awaited amplicon specific to E. histolytica. The absence of gene amplification for the third sample could be due to either degraded quality or insufficient amount of specific DNA.
Discussion
The formalin fixed embedded tissues constitute a source of genetic material widely used in many fields of research and in the diagnosis of many diseases (Dedhia et al., 2007). They are adapted to many retrospective studies thanks to the huge available collections of such biological material that is easier to conserve compared to fresh or frozen tissues (Weiss et al., 2011). However, the extracted DNA from FFPET is often degraded with poor quality namely because of the fixing process and the long duration of storage (Greer et al., 1991). The relevance of the dewaxing and extracting DNA methods used is crucial and was consequently largely discussed (Rivero et al., 2006; Funabashi et al., 2012; Weiss et al., 2011; Farrugia et al., 2010; Cao et al., 2003; Shi et al., 2002; Chan et al., 2001; Howe et al., 1997; Coombs et al., 1999). The current study aimed to optimize a simple, reliable, cost effective and efficient extraction method.
The results did not show any statistical difference between the two tested dewaxing techniques (xylene and temperature). Many authors stress the importance of this step by influencing the extraction efficiency (Piniewska et al., 2012; Santos et al., 2009) as well as in reducing potential PCR inhibitors (Stanta and Schneider, 1991). On the other hand, some authors argue that this treatment is unnecessary and do not improve the amount of DNA extracted and the success of the PCR amplification (Gilbert et al., 2007).
All tested combinations provided good amounts of DNA excluding xylene/salting-out combination which was significantly less efficient (p=0.001). The commercial kit (Qiagen) was associated to highest DNA purity (p<0.001).
The best success rates concerning PCR amplification of the IL6 human gene were obtained with the xylene/phenol-chloroform, xylene/kit and temperature/kit combinations (Table 2).
Although, efficient amount and purity of DNA were obtained from temperature/phenol-chloroform and temperature/salting-out combinations, only 30% of corresponding PCR results revealed positive. The degradation of DNA dosed using Nano Drop and the presence of PCR inhibitors may explain such situation. The temperature/Qiagen kit combination revealed the most efficient with good DNA yield and high purity. Moreover, this combination is easier to perform, faster, simple and does not use toxic substances. Similar conclusion was also reported by Funabashi et al. (2012). The positive amplifications of two out of the three tested cerebral amoebiasis FFPET submitted to temperature/Qiagen kit DNA extraction method highlight the efficiency of such combination also in microbiological DNA extraction. The commercial kits and particularly the QIAamp DNA Mini Kit (Qiagen) are widely used to extract DNA from FFPET (Funabashi et al., 2012; Dedhia et al., 2007; Farrugia et al., 2010). Carturan and his collaborators found, following a comparison of different commercial kits, that the Qiagen kit is efficient and adequate for FFPET (Carturan et al., 2008). However, the high cost of the kits is a limiting factor for their use in research and makes them dedicated to diagnosis purpose. The traditional phenol-chloroform extraction method was also effective while being cheaper. However, this method is laborious, time consuming (3 - 4 days), toxic and provides DNA contaminated with proteins. The simple, fast and non-toxic salting-out method was not retained because it provides DNA with lower degree of yield and purity (Table 1). However and unlike our findings, some authors have described this method as effective (Rivero et al., 2006; Howe et al., 1997; Liu and Zhang, 2011; Mirmomeni et al., 2010). The result obtained using this combination may be due to the persistence of the salt in the solution containing DNA and the degradation of the DNA.
Conclusion
Formalin fixed paraffin-embedded tissue represent an important source of DNA. The development of efficient protocol for extracting DNA from such sample is crucial. The combination temperature/commercial kit showed good results in preserving DNA quality and quantities.
Acknowledgment
We thank Mrs. Thelja Assili, Dr. Haifa Tounsi and Dr. Samir Boubaker from Laboratory of Pathology, Pasteur Institute of Tunis for their precious help.
Conflict of interest
The authors have declared no conflicting interests.
Authors’ Contribution
RJ, RBA and HC designed and performed the study, and wrote paper. NBA analyzed data, SH collected samples, KA and AB designed and supervised the study and corrected paper.All authors read and approved the final manuscript.
References





