Journal of Infection and Molecular Biology
Research Article
Prevalence of MRSA Containing SCCmec Types Isolated from a Tertiary Care Hospital, Rawalpindi
Muhammad Umer Arshad1*, Rao Waqas Akhtar2, Saira Irum3
1Rawalpindi Medical College, Rawalpindi; 2University of Health Sciences, Lahore; 3Quaid e Azam University, Islamabad, Pakistan.
Abstract | With the changes in epidemiology of methicillin-resistant Staphylococcus aureus (MRSA), accurate information on the scope and magnitude of MRSA infections is needed. This study was conducted to investigate the SCCmec types among MRSA strains collected from a hospital in Rawalpindi. Non-duplicate isolates of Staphylococci were isolated (n=28) from pus specimens (n=100). All strains were tested for methicillin resistance using Cefoxitin disc (30 µg) on Muller-Hinton agar. The phenotypically confirmed isolates (n=22) were screened for the presence of mecA, nuc genes and SCCmec types through multiplex PCR. Both mecA and nuc genes were detected in all Staphylococcus aureus strains, whereas, two SCCmec types III and I were detected in (77.27%) and (31.81%) of isolates, respectively along with subtypes. Antimicrobial susceptibility revealed Vancomycin as the most effective drug against all clinical isolates of MRSA. It is concluded that prevalence of MRSA is quite high in and around Rawalpindi area and thus requires necessary interventions for its effective treatment and control.
Keywords | Staphylococcus aureus, Infection, MRSA, mecA, nuc genes, SCCmec Types
Editor | Tahir Yaqub, University of Veterinary and Animal Sciences, Lahore, Pakistan.
Received | July 31, 2015; Revised | October 15, 2015; Accepted | October 21, 2015; Published | November 17, 2015
*Correspondence | Muhammad Umer Arshad, Rawalpindi Medical College, Rawalpindi, Pakistan; Email: umeruhs@gmail.com
Citation | Arshad MU, Akhtar RW, Irum S (2015). Prevalence of MRSA containing SCCmec types isolated from a tertiary care hospital, Rawalpindi. J. Inf. Mol. Biol. 3(4): 81-85.
DOI | http://dx.doi.org/10.14737/journal.jimb/2015/3.4.81.85
ISSN (Online) | 2307-5465; ISSN (Print) | 2307-5716
Copyright © 2015 Arshad et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Staphylococcus aureus isolates including methicillin resistant Staphylococcus aureus (MRSA) are the most common causes of skin and soft tissue infections, bacteremia, osteomyelitis and other invasive infections (David and Daum, 2013). MRSA is one of the nosocomial pathogen that cause about 20% of bloodstream infections (Klevens et al., 2007). These are also responsible for high morbidity and mortality and longer hospital stays (Park et al., 2013). With the acquisition of mecA gene they showed high resistance to all penicillins and cephalosporin’s (beta-lactams) (Lowy, 1998). The mecA gene is located on a mobile genomic island called Staphylococcal cassette chromosome mec (SCCmec) (Bode et al., 2012). Nuc gene is located on a plasmid (pIP1607) which encodes the thermo-stable nuclease of S. aureus (Brakstad et al., 1992). SCCmec elements are highly diverse in their structural organization and genetic content (Elements, 2009). There are total 11 SCCmec types but generally type IV and V are harbored by (HA-MRSA) healthcare-associated MRSA while type I, II and III is harbored by CA-MRSA (Community acquired MRSA) (Dhawan et al., 2014). SCCmec typing has become essential for the characterization of MRSA in epidemiological studies (Chongtrakool et al., 2006). Accurate and rapid identification of MRSA in clinical specimens is essential for effective antimicrobial chemotherapy (Jonas et al., 2002). The aim of this study was to determine the prevalent MRSA genotypes circulating in the Holy Family Hospital (HFH) Rawalpindi between November 2012 and January 2013 using PCR, SCCmec typing and antimicrobial susceptibility testing.
METHODOLOGY
A total of one hundred (n=100) pus specimens were collected from anterior nares and perirectal area of patients attending (RMC) Allied Hospitals, Rawalpindi aseptically. Among these (n=100) samples, (n=50) were collected from paeds intensive care unit (PICU) and (n=50) were from medical intensive care unit (MICU) collected during November 1, 2012 to 31th January 2013. The specimens were collected as described by (Allen 2001) and transported to the laboratory within an hour of collection for processing.
The samples were inoculated on Mannitol salt agar and incubated at 37°C aerobically for 24 h. The identification of Staphylococci was done by their cultural characteristics, colony morphology and their biochemical profile (catalase test, coagulase test and DNAse test) following the scheme of (Barrow and Feltham, 2004). 30ug Cefoxitin disc was used to confirm Staphylococci as MRSA as per CLSI guidelines 2012.
DNAs were extracted from MRSA strains for the amplification of SCCmec types, mecA and nuc genes by ethanol precipitation method. Both the mecA and nuc genes were detected using single targeted primers in multiplex PCR as already described in literature (Murakami et al., 1991; Dhawan et al., 2014). The amplicon were visualized under UV light on 1% agarose gel electrophoresis.
SCCmec type genes were detected using single targeted primers in multiplex PCR as already described in literature (Oliveira and de Lencastre, 2002). The Amplicon were visualized under UV light on 1% agarose gel electrophoresis.
Antimicrobial susceptibility testing of Staphylococci was performed by Kirby-Bauer disc diffusion method on Mueller-Hinton agar as per CLSI guidelines 2012. Implanted antibiotics were Penicillin (10 µg), Methicillin (30 µg), Oxacillin (5 µg), Amoxicillin (30 µg), Cephalaxin (30 µg), Cefoxitin (30 µg), Cephalothin (30 µg), Cephradine (5 µg), Ciprofloxacin (5 µg), Gentamicin (30 µg), Imipenum (10 µg), Levofloxacin (5 µg), Tetracycline (30 µg), and Vancomycin (30 µg). Plates were incubated at 37°C overnight aerobically.
The interpretation was done as per CLSI guidelines. Statistical analysis was done using SPSS 16.0.
RESULTS
Out of 100 pus specimens, only 28 were identified as Staphylococci. Among these, 22 were Staphylococcus aureus (coagulase positive) and 6 were coagulase negative Staphylococci (CoNs) (n=6). On the basis of Cefoxitin disc, 22 Staphylococci were identified as MRSA.
PCR was carried out with DNA extracted from all MRSA strains and control strain S. aureus N315 was used as a template. The expected band of 533bp for mecA and 278bp for nuc genes were amplified in all phenotypically confirmed MRSA strains by the PCR amplification method (Figure 1).
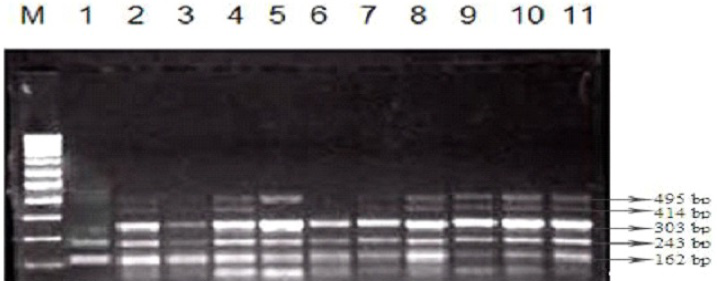
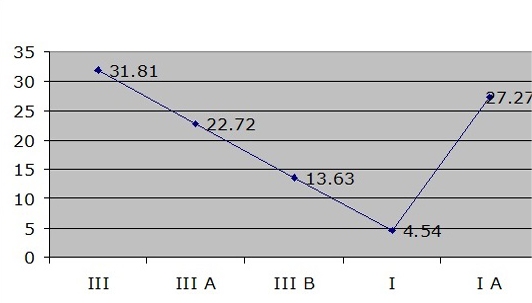
Out of 22 strains, only 7 strains were positive for SCCmec type I while 17 strains were positive for SCCmec type III. A clear and easily discriminated band pattern was obtained for the required SCCmec types using the multiplex PCR. The most prevalent SCCmec type was found to be type III (31.81%) then IA (27.27%) followed by III A (22.72%) and III B (13.63%) and least prevalent type was SCCmec type I (4.54%) (Figure 4).
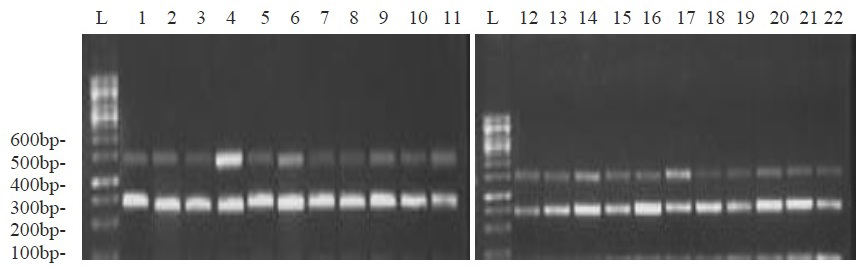
Figure 1: Multiplex PCR based electrophoresis pattern of isolates for nuc (278bp) and mecA gene (533bp)
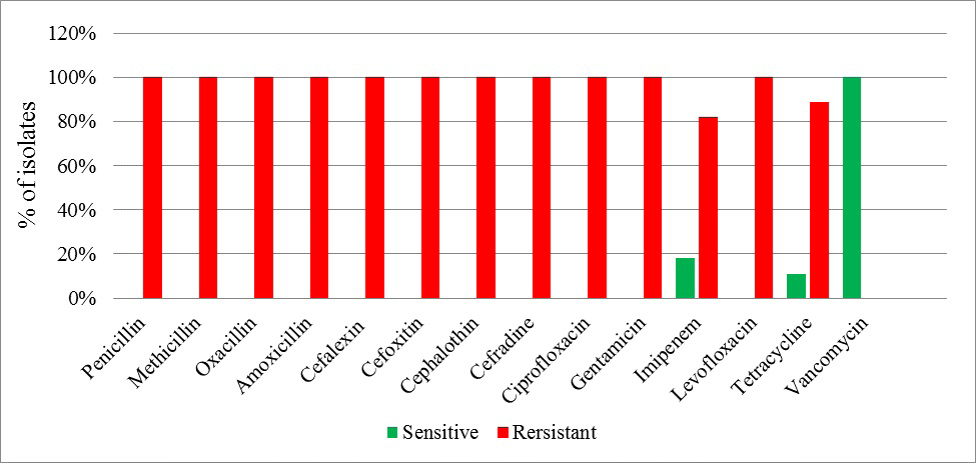
Figure 2: Antimicrobial resistance profile of study isolates against the common antibiotics used in treatment
The two major SCCmec types along with subtypes were easily distinguishable: type I strains displayed a band of 495bp and type III strains display two bands of 414bp and 243bp (Figure 3).
Overall the Staphylococci showed 100 % resistance against commonly used antibiotics while most effective drug was Vancomycin (Figure 2).
Table 1: Specific primers used for mecA and nuc genes
|
Primer |
Nucleotide Sequences |
Product size |
|
mecA (F) |
AAA-ATC-GAT-GGT-AAA-GGT-TGG-C |
533 bp |
|
mecA (R) |
AGT-TCT-GCA-GTA-CCG-GAT-TTG-C |
|
|
nucA (F) |
GCG-ATT-GAT-GGT-GAT-ACG-GTT |
278 bp |
|
nucA (R) |
AGC-CAA-GCC-TTG-ACG-AAC-TAA-AGC |
DISCUSSION
Approximately 20% of healthy persons are persistent carriers of S. aureus, and 80% are intermittent carriers. Colonization rates are increased in hemodialysis patients, illicit injection drug users, surgical patients and patients with insulin dependence or poorly controlled diabetes (Moreillon et al., 2005). MRSA is often the severe causal agent in nosocomial infections that are becoming increasingly difficult to cure because of emerging resistance to all current antibiotic classes (David and Daum, 2010). Novel MRSA clones keep occurring in hospitals and more recently in the community often causing sustained epidemics. A survey report documented the prevalence of MRSA from 42% to 51% in Pakistan (Sieradzki et al., 1999; Parola et al., 2004).
Risk factors associated with acquiring MRSA in hospitals includes prolonged hospitalization, preceding antimicrobial therapy, surgical procedures and proximity to another patient known to be colonized or infected with MRSA. The global emergence of different clones of methicillin-resistant Staphylococcus aureus (MRSA) has arguably been the major hindrance in the history of antimicrobial therapy (Gould, 2009). The reason for development of antibiotic resistance in developing countries like ours seems to be very much related to the irrational antibiotic usage due to its easy availability at the drug store without prescription.
In present study the state of resistance exists among clinical isolates of S. aureus obtained from ICUs of tertiary health care facilities were explored. We also determined the prevalence of MRSA a superbug of hospital care infections and also set up a rapid and reliable identification procedure for MRSA by the amplification of specific gene determinants like nuc and mecA genes by PCR. We also characterize isolated MRSA with SCCmec typing by help of multiplex PCR.
Prevalence of MRSA varies widely not only in Pakistan but also in other countries. In current study, the prevalence of MRSA among total isolates was found to be 22% comparable to (Hafiz et al., 2002) and (Bukhari et al., 2004) who reported 42% and 38.6%, respectively. Various other studies also showed very high prevalence i.e. 65% by (Safdar et al., 2003). The detection of mecA gene which is responsible for the resistance of S. aureus to methicillin was considered as gold standard for confirmation of MRSA in isolated samples (Skov et al., 2006). In present study we made mecA gene detection by PCR using primer of 533bp fragment and our results showed almost 99%, reliability.
The routine consignment of MRSA isolates to SCCmec types now practicable with the SCCmec multiplex PCR typing. It may researchers with important information concerning the origin of MRSA clones and the evolutionary relationships among these. SCCmec typing is essential because it can differentiate between HA-MRSA and CA-MRSA. As a result of different antibiotic susceptibility profiles between the HA and CA-MRSA, the discrimination of these two strains of MRSA would help in patient and antibiotic management (Kilic et al., 2008). In our study SCCmec typing by multiplex PCR showed that SCCmec
Table 2: Specific primers used in multiplex PCR for SCC mec typing
|
Primers Oligonucleotide sequence (5’–3’) |
Amplicon size (bp) |
Specificity(SCCmec type) |
|
TTCGAGTTGCTGATGAAGAAGG ATTTACCACAAGGACTACCAGC |
495 |
I |
|
AATCATCTGCCATTGGTGATGC CGAATGAAGTGAAAGAAAGTGG |
284 |
II |
|
ATCAAGACTTGCATTCAGGC GCGGTTTCAATTCACTTGTC |
209 |
II, III |
|
CATCCTATGATAGCTTGGTCCTAAATCATAGCCATGACCG |
342 |
I, II, IV |
|
GTGATTGTTCGAGATATGTGGCGCTTTATCTGTATCTATCGC |
243 |
III |
|
TTCTTAAGTACACGCTGAATCGGTCACAGTAATTCCATCAATGC |
414 |
III |
|
TCCAGATTACAACTTCACCAGG CCACTTCATATCTTGTAACG |
162 |
Internal control |
type III element (77.27%) was the predominant type followed by SCCmec type I (31.81%). All the SCCmec types identified in our study belonged to (HA-MRSA) MRSA strains. There is no difference in prevalence of the SCCmec genes isolated in samples from PICU and MICU.
In the present study all MRSA on disc diffusion method were found multiple drug resistant i.e. 100% resistance found against Penicillin, Methicillin, Oxacillin, Amoxicillin, Cephalaxin, Cefoxitin, Cephalothin, Cephradine, Ciprofloxacin, Gentamicin, Imipenum, Levofloxacin and Tetracycline. Vancomycin was found to be the most effective drugs against the MRSA isolates among the routinely used antibiotics. There are also reports from many other countries of isolates with reduced susceptibility to vancomycin and teicoplanin frequently associated with a significant thickening of the cell wall (Dyke, 2003). The emergence of vancomycin resistance in Pakistan is an alarming sign as 4% resistance was reported by (Bukhari et al., 2004) but still it remained an effective antibiotic against MRSA (Dar et al., 2006). But in our study there were no VRSA and VISA strains isolated among MRSA isolates.
The conclusions drawn from this study were that prevalence of MRSA in ICUs of tertiary health care facilities was found to be quite high (22%) resulting in increased incidence of MRSA infections there. So there is a need to take steps for effective eradication and treatment of MRSA infections. It is recommended that we should develop a rapid and reliable identification and antimicrobial susceptibility in order to efficiently support therapy, eradication of the pathogen and reduction in the costs associated with falsely diagnosed MRSA infection. Furthermore establishment of a molecular diagnostic technique PCR for detecting mecA in Staphylococci collected at tertiary care units of the country is also an important recommendation of the current study.
Conflict of interest
The authors declare that there is no conflict of interest regarding this study.
Authors’ Contribution
Irum S designed the study and planned research methodology. Akhtar RW contributed in drafting the article and revising it critically for important intellectual content. Arshad MU gave final approval of the version to be published.
Acknowledgements
We are extremely thankful to Dr. Arafat Yameen (COMSATS Institute of Information Technology, Abbottabad), Ms. Shaista Kanwal (Department of Microbiology QAU, Islamabad) and Abdul Basit (RMC, Rawalpindi) for their valuable technical assistance and unconditional support.
REFRENCES