Research Journal for Veterinary Practitioners
Research Article
Research Journal for Veterinary Practitioners 2(1): 13 – 16Antimicrobial Resistance Pattern against Staphylococcus aureus in Environmental Effluents
Muhammad Ahaduzzaman1, Mohammad Mahmudul Hassan1*, Mahabub Alam1, SKM Azizul Islam1, Inkeyas Uddin2
- Department of Physiology, Biochemistry and Pharmacology, Faculty of Veterinary Medicine, Chittagong Veterinary and Animal Sciences University, Khulshi, Chittagong–4225, Bangladesh
- Poultry Research and Training centre, Chittagong Veterinary and Animal Sciences University, Khulshi, Chittagong–4225, Bangladesh
*Corresponding author: miladhasan@yahoo.com
ARTICLE CITATION:
Ahaduzzaman M, Hassan MM, Alam M, Islam SKMA, Uddin I. (2014). Antimicrobial resistance pattern against Staphylococcus aureus in environmental effluents. Res. j. vet. pract. 2(1): 13 – 16.
Received: 2013–12–12, Revised: 2013–12–20, Accepted: 2013–12–21
The electronic version of this article is the complete one and can be found online at
(http://dx.doi.org/10.14737/journal.rjvp/2014/2.1.13.16)
which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
Abstract
Effluents from hospitals and slaughterhouses are alarming threat for clinician to treat the patient with antibiotics due to harboring resistance bacteria. The study was undertaken to evaluate the antimicrobial resistance pattern against Staphylococcus aureus in hospital and slaughterhouse effluents. Staphylococcus aureus from six medical hospitals, five veterinary hospitals and five slaughter houses were isolated by using different methods. This Staphylococcus aureus isolates was used to find out the antibiotic resistance pattern by using disc diffusion method. The antibiotic resistance patterns of identified isolates showed that Amoxicillin, Cefradin, Colistin, Cefalexin, Oxytetracycline and Pefloxacin were 100% resistance to hospitals and slaughterhouses isolates (except colistin 75% in slaughterhouse), and Enrofloxacin were 80%, 50%, 75%; Gentamicin were 40%, 50%, 50%; Kanamycin were 40%, 50%, 75% and Neomycin were 40%, 50% and 25% respectively, resistance to medical hospitals, veterinary hospitals and slaughterhouses isolates. Results indicated that hospitals and slaughter houses waste effluent has multiple–antibiotic resistance Staphylococcus aureus. Based on this study, some efforts have to be taken to reduce the possibility of resistant bacteria entering into and spread in the environments for securing possible public health threat.
INTRODUCTION
Staphylococcus aureus is one of the ubiquitous microorganisms and are frequently associated with various infections. Antibiotic resistant Staphylococcus aureus isolates pose an unbelievable challenge to both veterinary and human health professionals because they have a pessimistic therapeutic impact (Brouillette and Malouin, 2005). The indiscriminate usage of antibiotics correlates with the emergence, maintenance and transmission of antibiotic resistant traits within pathogenic strains (Shitandi and Sternesjö, 2004). These resistance traits are coded by particular genes that may be carried on the bacterial chromosome, plasmids, on gene cassettes that are incorporated into integrons (Rychlik et al., 2006), thus are transferred among isolates an easy manner. National estimates from 2001 to 2002 in USA suggest that 32.4% of individuals are colonized with Staphylococcus aureus, and 0.8% of individuals were colonized with MRSA (Graham et al., 2006). Biomedical wastes pose great danger in Bangladesh as estimated that 20% of the biomedical waste is “highly infectious” and is a hazard since it is often disposed of into the sewage system, drains or water bodies. Such poor sanitation has serious consequences for the health of the residents and a report suggests that “most of the child mortality” could be related with this problem (Bhuiya, 2007). Resistance bacterial contamination of waste waters worldwide is estimated to cause millions of gastrointestinal and acute respiratory infections (Shuval, 2003) and numerous skin infections (Yau et al., 2009) every year. Unlike typical hospital associated strains, some strains of community associated Staphylococcus aureus can cause infections in healthy people with no traditional risk factors for infection (Gorwitz, 2008). Even though most patients are treated as outpatients, hospitalization rates remain substantial (Jarvis et al., 2007). Similarly, abattoirs are generally known all over the world to pollute the environment either directly or indirectly from their various processes (Adelegan, 2002). Health risks of the effluents of slaughterhouses play role in environment degradation and to dissemination of pathogenic resistance microorganisms during the rainy and dry seasons which not manage properly. Such a practice has led to misuse of antimicrobial drugs with the associated high prevalence of drug resistance among the staphylococci (Okuma, 2002). Multiple antibiotic resistant Staphylococcus aureus strains have been isolated from milk, meat, water, human and environmental samples in many parts of the world (Pesavento et al., 2007) but minimal concern gather towards multidrug resistance Staphylococcus aureus isolates from effluents in Indian subcontinent. A majority of the prescribers in Bangladesh particularly in veterinary practice made diagnosis of infection based on clinical assessment and suspect a microbial etiology due to inadequate diagnostic facilities and frequently choose higher antibiotics (Faiz and Rahman, 2004). The important factors associated with resistant bacteria with poor hospital hygiene, overcrowding, lack of resources for infection control and lack of personnel trained in controlling infection in both medical and veterinary hospital (Faiz and Basher, 2011).
Bangladesh is the populous country having its place at ninth most densely populated countries in the world. In particular, from 2010–2015 the projected urban population growth rate is 3% (UNdata, 2012). With this population growth, there is an increasing problem of waste management particularly in the larger cities due to inadequate budget and awareness. The minimal waste collection coverage forces majority of the waste to be dumped in open lands or drainage system. These wastes are not disposed properly, where general wastes are often mixed with hazardous waste such as hospital waste, slaughterhouse waste and other environmental wastes. Frequently these wastages clog together due to solid waste and submerge the public place and cultivable land and contaminated the surroundings with resistance microbes. Therefore the current studies were taken out to isolates the Staphylococcus aureus and find out their antibiotic resistance pattern from environmental effluents.
MATERIALS AND METHODS
Study Area
The study was carried out in Chittagong Metropolitan area had an estimated population of more than 6.5 million people. At present government, non–government hospitals and clinics have provided to the medical service for the city civilian and also a number of veterinary hospitals are present for animal health care. On the other hand, some slaughterhouses are also established scattered within the city. Six (6) medical hospitals, five (5) veterinary hospitals and five (5) slaughterhouses were selected randomly from the list of Chittagong City Corporation.
Study Duration and Sample Collection
The study was conducted during the period of September to December, 2012. Samples were collected from final effluents of medical hospitals, veterinary hospitals and slaughterhouses and 250ml pre–sterilized glass bottles were used to transport the samples to PRTC (Poultry Research and Training Center) laboratory for analysis.
The collected samples were preserved in a refrigerator chiller at 40C during the research period at PRTC laboratory.
Media Used
Nutrient broth (Oxoid Ltd., PH: 7.4±0.2) was used as primary enrichment media for Staphylococcus. Selective media Mannitol salt agar (Merck, PH: 7.4±0.2) was used for isolation of the bacteria and Muller Hinton agar (Biotec, PH: 7.3±0.1) used for determination of antibiogram.
Isolation and Identification of Organism
For the isolation of Staphylococcus aureus, 1ml of water sample was inoculated in screw cap test tube containing nutrient broth (primary enrichment media) and incubated overnight at 370C. After primary enrichment sample from nutrient agar was streaked on mannitol salt agar and incubated for 24 hours at 370C. After overnight incubation the bacterial growth was observed and the positive samples colony were selected based on colony morphology and confirmed by Gram’s staining.
Gram Staining
Gram’s staining was performed as per procedures described by Merchant and Packer (1969) to determine the size, shape and arrangement of bacteria.
Catalase Test
Pure culture of each isolate was picked using a sterile loop from the agar slant and mixed with a drop of 3% H2O2 on a clean glass slide. Bubbles of oxygen were liberated within a few seconds.
Coagulase Test
Slide Coagulase Test
Dense suspensions of Staphylococci from culture were made on two ends of clean glass slide. One was labeled as “test” and the other as “control”. The control suspension serves to rule out false positivity due to auto agglutination. The test suspension was treated with a drop of EDTA treated horse plasma and mixed well. Agglutination or clumping of cocci within 5–10 seconds was taken as positive. Some strains of Staphylococcus aureus may not produce bound coagulase, and such strains must be identified by tube coagulase test.
Tube Coagulate Test
The tube Coagulase test was performed in sterile tubes by adding 0.5ml of isolates of Staphylococcus grown on tryptone soya broth (TSB) at 37°C for 24h to 0.5ml of Goat plasma. After mixing by gentle rotation, the tubes were incubated at 37°C along with a negative control tube containing was evaluated at 30 minutes intervals for the first 4h of the test and then after 24h incubation. The reaction was considered was positive if any degree of clotting from a loose clot to a solid clot that is immovable when the tube is inverted (tilted) was visible within the tube and no degree of clotting would be taken as negative.
CS test at Muller Hinton Agar
After confirmation of isolates as Staphylococcus aureus, antimicrobial susceptibility of the isolates were determined by using the micro disc diffusion method, and the method was used according to guidelines established by Clinical and Laboratory Standards Institute (CLSI) (2010).
Data Analysis
Data obtained was imported to the Microsoft Office Excel–2007 and transferred to the software STATA/IC–11 for analysis. Descriptive statistics was done and expressed as percentages of resistance pattern of antimicrobials.
RESULTS
The results of isolation and identification of Staphylococcus aureus in culture of Mannitol salt agar, Gram staining, Catalase and Coagulase test were given in table–1. The Staphylococcus aureus ferments mannitol and turned the medium yellow. The positives isolated from mannitol salt agar microscopically detected as Gram–positive, coccid as clustered of grapes. The colonies were characterized as round, smooth and glistening that was positive in 5 Medical hospital, 2 Veterinary hospital and 4 slaughterhouse samples, as mannitol salt agar is selective media for Staphylococcus spp. Catalase and Coagulase test was done for conformation of Staphylococcus aureus and find all the sample positive.
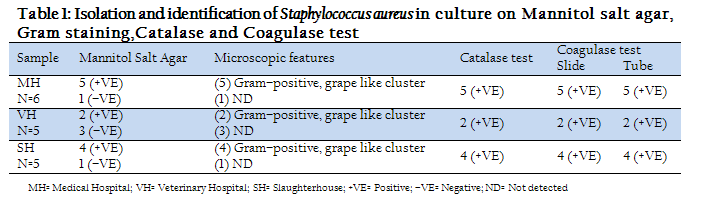
Table 1: Isolation and identification of Staphylococcus aureus in culture on Mannitol salt agar, Gram staining,Catalase and Coagulase test
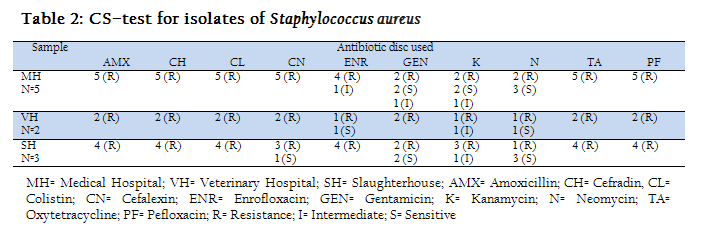
Table–2 represents the results of CS–test for isolates of Staphylococcus aureus. Ten antibiotics were used from which AMX, CH, CL, CN, TA and PF were resistance to all five medical hospitals isolates. ENR, GEN and K were intermediate sensitive to 1 medical hospital isolates but shown resistance to rest of the three. GEN, K and N were sensitive to medical hospital isolates. AMX, CH, CL, CN, TA and PF were resistance to all Veterinary hospital isolates, K was intermediate sensitive and ENR, GEN and N were sensitive to one isolates. All Staphylococcus aureus isolates from slaughterhouses were able to shown resistance against AMX, CH, CL, TA, ENR and PF. Sensitivity to CN was only found in one sample and was resistance to others.
The prevalence of antibiotic resistance pattern against Staphylococcus aureus positive isolates were shown in table–3. The percents of resistance among antibiotics were 100% to AMX, CH, CL, CN, PF and TA followed by ENR (80%), GEN (40%), K (40%) and N (40%) in the isolates of medical hospital. The prevalence of antimicrobial resistance pattern against Staphylococcus aureus were 100% to AMX, CH, CL, CN, TA and PF followed by ENR (50%), GN (50%), K (50%), N (50%) in the isolates of veterinary hospital. The level of resistance for slaughterhouse Staphylococcus aureus isolates to specific antibiotics were 100% to AMX, CH, CN, TA and PF followed by 75% resistance in CL, ENR and K, 50% to Gentamicin and 25% to N.
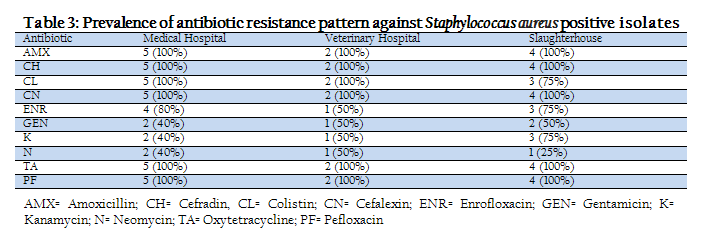
Table 3: Prevalence of antibiotic resistance pattern against Staphylococcus aureus positive isolates
DISCUSSION
Staphylococcus aureus, a spherical, aerobic, Gram positive, catalase positive, coagulase positive, is an opportunistic pathogen for both human and livestock population, and is one of the most frequent etiological sources of hospital and community infections. Generally, Staphylococcus aureus is responsible for superficial pyogenic infections as abscess, systemic infections such as endocarditis, osteomyelitis and pneumonia, and toxicosis such as food poisoning or toxic shock syndrome after ingestion of contaminated food. The isolates of Staphylococcus aureus from medical hospitals, veterinary hospitals and slaughterhouses were tested to find out the antimicrobial resistance pattern. The effluents were contaminated by the large number of patients in both human and animal hospitals, and different species of animals in slaughterhouses, and high possibilities of contamination of existent Staphylococcus aureus in the effluents of those places. Antimicrobial resistance pattern of the Staphylococcus aureus from samples of hospitals and slaughterhouses showed high resistance levels. The level of resistance is higher than the findings described by Virdis et al. (2010) and that might be due to variation of environment and contaminated effluents. A study reported that, Staphylococcus resistance to oxacillin, penicillin and ampicillin was 100% and cephalothin was 92.4% (Bukhari et al., 2011) and those were agreed to present findings. Kanamycin resistance was not acute in our present research and effectiveness was still showed against hospital isolates but minor sensitivity was showed against slaughterhouse isolates. Our results support the findings of the prevalence of kanamycin resistance bacteria in drinking water from a residential area and closed to the values of resistance for isolates from college drinking water but differ from the values obtained from hospital drinking water (Samra et al., 2009). The variation in prevalence of antimicrobial resistance patterns might be due to differences in hygienic condition and contamination of water in study area.
Chu et al. (2012) reported 137 staphylococcal strains were identified predominantly from milk, and in the vagina, anus, and nasal cavity. Nearly all isolates were found susceptible to enrofloxacin, gentamicin, neomycin, and vancomycin and in addition, over 60% of the isolates were also resistant to penicillin, ampicillin, cephalothin, tetracycline, and oxytetracycline that support our findings. Multidrug resistance is a great problem in the therapeutic grounds especially in third world countries due to indiscriminate use of antimicrobials and not maintaining proper withdrawal period. Watéba et al. (2013) found MRSA isolates were resistant to kanamycin (88%), tobramycin (82%), gentamicin (64%), and pefloxacin (70%) which partially support our findings. There is tremendous rate of waste generation in Bangladesh and has possibilities to reach 47,064 tons per day by 2025 and the effluent generation rate (kg/cap/day) is expected to increase to 0.6 in 2025 (Alamgir and Ahsan, 2007). A significant percentage of the population has zero access to proper waste disposal knowledge and practice, which will in effect lead to the problem of waste mismanagement and antibiotic resistance. Some efforts have to be made to reduce the possibility of resistant bacteria entering into and spread in the environments. The discharge of wastewater containing resistant bacteria in the environment poses a real public health problem; hence the importance of the installation and operation of treatment plants to reduce the rate of waterborne diseases have to be taken. The importance of the different sources of resistance found in the environment, i.e. the presence of antibiotics in the environment and the importance of resistant bacteria resulting from the use of antibiotics in the various fields has to be measured. For this purpose, it is important to make a more detailed assessment of the significance of culture–dependent and laboratory–based methods in relation to conditions found in the environment. Based on the above resistance pattern of antimicrobial agents on environmental samples, measures should be taken to avoid or control the antimicrobial resistance and improve the public health.
ACKNOWLEDGEMENTS
Authors are grateful to the Poultry Research and Training Centre (PRTC), Chittagong Veterinary and Animal Sciences University, Bangladesh for giving laboratory support to the present research project.
COMPETING INTERESTS
Authors declare that they have no competing interests.
REFERENCES
Adelegan JA (2002). Environmental Policy and Slaughterhouse waste in Nigeria, Proceedings of the 28th WEDC Conference Kolkata (Calcutta), 3–6.
Alamgir M and Ahsan A (2007). Municipal solid waste and recovery potential: Bangladesh perspective. Iranian. J. Environ. Health. Sci. Eng. 4(2):67–76.
Bhuiya GMJA (2007). Bangladesh solid waste management. Edited by the Environmental Management Centre, Mumbai, India, 30.
Brouillette E and Malouin F (2005). The pathogenesis and control of Staphylococcus aureus induced mastitis: Study models in the mouse. Microbes. Infect. 7:560–568.
http://dx.doi.org/10.1016/j.micinf.2004.11.008
PMid:15777742
Bukhari SZ, Ahmed S and Zia N (2011). Antimicrobial susceptibility pattern of Staphylococcus aureus on clinical isolates and efficacy of laboratory tests to diagnose MRSA: a multi–centre study. J. Ayub. Med. College., 23:139–42.
Chu C, Yu C, Lee Y and Su Y (2012). Genetically divergent methicillin–resistant Staphylococcus aureus and sec–dependent mastitis of dairy goats in Taiwan. BMC Vet. Res. 8:39.
http://dx.doi.org/10.1186/1746-6148-8-39
PMid:22455622
CLSI (Clinical and Laboratory Standards Institute) (2010). Changes in the CLSI standards for antimicrobial susceptibility testing for 2010. (http://www.apicchicago.org/pdf/2010).
Faiz MA and Basher A (2011). Antimicrobial resistance: Bangladesh experience. Regional Health Forum, 15 (1): 1–8.
Faiz MA and Rahman MR (2004). Rational antimicrobial use. J. Chittagong. Med. College. Teacher. Associ. 15(1-2): 1–3.
Gorwitz RJ (2008). A review of community associated methicillin–resistant Staphylococcus aureus skin and soft tissue infections. Pediatr. Infect. Dis. J. 27(1):1–7.
http://dx.doi.org/10.1097/INF.0b013e31815819bb
PMid:18162929
Graham PL, Lin SX and Larson EL (2006). A U.S. population–based survey of Staphylococcus aureus colonization. Ann. Intern. Med. 144:318–325.
http://dx.doi.org/10.7326/0003-4819-144-5-200603070-00006
PMid:16520472
Jarvis WR, Schlosser J, Chinn RY, Tweeten S and Jackson M (2007). National prevalence of methicillin–resistant Staphylococcus aureus in inpatients at US health care facilities, 2006. Am. J. Infect. Control. 35(10):631–637.
http://dx.doi.org/10.1016/j.ajic.2007.10.009
PMid:18063126
Merchant IA and Packer RA (1969). Veterinary Bacteriology and Virology. 7th edn., The Iowa State University Press, Ames, Iowa, USA. 211–305.
Okuma K, Iwakawa K, Turnidge J, Grubb WB, Bell JM, O'Brien FG, Coombs GW, Pearman, JW, Fred C, Tenover FC, Kapi M, Tiensasitorn C, Ito T and Hiramatsu K (2002). Dissemination of new methicillin– resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289–4294.
http://dx.doi.org/10.1128/JCM.40.11.4289-4294.2002
PMid:12409412 PMCid:PMC139674
Pesavento G, Ducci B, Comodo N and LoNostro A (2007). Antimicrobial resistance profile of Staphylococcus aureus isolated from raw meat: A research for methicillin resistant Staphylococcus aureus (MRSA). Food. Control. 18(3):196–200.
http://dx.doi.org/10.1016/j.foodcont.2005.09.013
Rychlik I, Gregorova D and Hradecka H (2006). Distribution and function of plasmids in Salmonella enterica. Vet. Microbiol. 112(1):1–10.
http://dx.doi.org/10.1016/j.vetmic.2005.10.030
PMid:16303262
Samra ZQ, Naseem M, Khan SJ, Dar N and Athar MA (2009). PCR Targeting of Antibiotic Resistant Bacteria in Public Drinking Water of Lahore Metropolitan, Pakistan. Biomed. Environ. Sci. 22, 458–463.
http://dx.doi.org/10.1016/S0895-3988(10)60002-5
Shitandi A and Sternesjö A (2004). Prevalence of multidrug resistant Staphylococcus aureus in milk from large and small–scale producers in Kenya. J. Dairy. Sci. 87:4145–4149.
http://dx.doi.org/10.3168/jds.S0022-0302(04)73557-2
Shuval H (2003). Estimating the global burden of thalassogenic diseases: human infectious diseases caused by wastewater pollution of the marine environment. J. Water. Health. 1(2): 53–64.
PMid:15382734
UNdata (2012). Country Profile: Bangladesh
Virdis S, Scarano C, Cossu F, Spanu V, Spanu C and DeSantis EPL (2010). Antibiotic resistance in Staphylococcus aureus and coagulase negative Staphylococci isolated from goats with subclinical mastitis. Vet. Med. International. (517060):1–6.
http://dx.doi.org/10.4061/2010/517060
PMid:20445785 PMCid:PMC2860459
Watéba I, Salou M, Ekouevi D, Dosseh D, Dossim S, Tigossou SD, Dagnra and AY Prince–David M (2013). P193: Nasal carriage of methicillin resistant Staphylococcus aureus in staff of the surgical services of CHU Sylvanus Olympio Lome–Togo. Antimicrob. Resist. Infect. Control. 2(1): 93.
Yau V, Wade TJ, DeWilde CK and Colford JM (2009). Skin related symptoms following exposure to recreational water: a systematic review and meta–analysis. Water. Quality. Expos. Health. 1:79–103