Journal of Animal Health and Production
Research Article
Journal of Animal Health and Production 1 (2): 20–23Characterization and Antimicrobial Sensitivity of Staphylococcus aureus Isolates from Subclinical Bovine Mastitis
Sant Prasad Tyagi, Rajesh Kumar Joshi*, Namita Joshi
*Corresponding author:rkjoshivet26@gmail.com
ARTICLE CITATION:
Tyagi SP, Joshi RK, Joshi N (2013). Characterization and Antimicrobial Sensitivity of Staphylococcus aureus Isolates from Subclinical Bovine Mastitis. J Anim. Health Prod. 1(2):20–23.
Received: 2013–04–12, Revised: 2013–06–25, Accepted: 2013–06–26
The electronic version of this article is the complete one and can be found online at
(http://nexusacademicpublishers.com/table_contents_detail/11/54/html)
which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
Abstract
Pathogenic Staphylococcus aureus was isolated from the cases of subclinical mastitis in lactating cows. A total of 214 quarter milk samples were screened of which, 110 (51.40%) samples were found positive for subclinical mastitis (SCM) using California Mastitis test (CMT). Isolation of Staph. aureus was attempted in 68 samples, comprising 52 distinct and 16 strong CMT positive samples. All 68 samples yielded Staph. aureus. All the isolates were catalase positive while 54 (79.41%) and 56 (82.35%) isolates showed positive slide and tube coagulase test, respectively. On sugar fermentation all the isolates were positive for mannitol fermentation, while 59 (86.76%) and 53 (77.94%) isolates fermented glucose and lactose. On sheep blood agar, 48 (17.58%) isolates showed β–haemolysis and 59 (86.76%) samples were positive for nitrate reduction test. Coa gene was detected in all the Staph. aureus while 22 (32.35%) isolates revealed Spa (X region) gene. All the isolates were sensitive to Cefotaxime while highest number of isolates 63 (92.64%), were found resistant to ampicillin, followed by 20 (29.41%) for tetracycline, 17 (25.00%) for ofloxacin, 11 (16.17%) for lincomycin and 09 (13.23%) for ciprofloxacin, respectively.
INTRODUCTION
Mastitis is single largest problem in dairy animals causing economic losses in the tune of Millions of rupees annually in India. Although, Mastitis is a multietiological disease, Staphylococcus aureus is the primary and probably the most lethal agent that causes chronic and deep infection in the mammary glands which is extremely difficult to be cured (Saravanan et al., 2000; Gianeechini et al., 2002; Wani et al., 2003). Rapid identification of such pathogenic bacteria can significantly reduce the time and cost of testing, resulting into timely cure, low cost of treatment and can significantly reduce production losses. The traditional method of bacterial culture is labor intensive and time consuming. Therefore, rapid PCR based methods have been developed to identify real pathogens. Although, very few strains of Staph. aureus do not produce detectable amount of coagulase, all strains seem to possess a coagulase gene (Coa) (Vandenesch et al., 1994). Similarly protein A, which is the major surface proteins of staphylococci (Spa), binds immunoglobulin G and impairs opsonisation by serum complement and phagocytosis by polymorphonuclear leukocytes (Gao and Stewart, 2004). Development of PCR–based methods for detection of such genes enabled rapid identification of virulent Staph. aureus (Riffon et al., 2001; Strommenger et al., 2006). The single most common use of antimicrobial agents in dairy cattle is for prevention and treatment of bovine mastitis (Kaneene and Miller, 1992; Moore and Heider, 1984). The resistance of Staph aureus to antimicrobial agents has been extensively documented and it contributed significantly to the treatment failure ( Sumathi et al., 2008; Sudhakar et al., 2009; Kumar et al., 2010). Present investigation reports the isolation and identification of pathogenic Staphylococcus aureus in the subclinically mastitic milk samples and their antibiogram.
MATERIALS AND METHODS
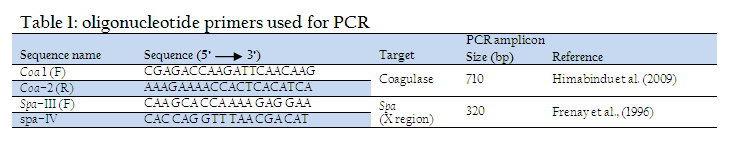
A total of 214 quarter milk samples were collected from lactating cows as per the recommendations of National Mastitis Council (NMC, 1990), transported on ice to Microbiology laboratory for further processing. The samples were subjected to California mastitis test (CMT) according to the procedure described by Quinn et al. (1994) and were graded as negative, trace(+1), weak(+2), distinct (+3), or strongly positive (+4). A total of 68 milk samples, graded as distinct and strongly positive, were selected for isolation of Staph. aureus using Mannitol salt agar as selective media. The isolates were characterized by Gram’s staining, haemolysis on 7% sheep blood agar and biochemical tests viz. Catalase test, Coagulase test (bound as well as free coagulase), nitrate test and fermentation of glucose, mannitol and maltose sugars (NMC, 1990, Quinn et al., 1994). The isolates were tested for the presence of coagulase (Coa) and Protein A (Spa x region) genes specific for virulent Staph. aureus using PCR. The heating technique described by Franco et al. (2008) was used for preparation of DNA template. The isolates were grown overnight at 370C in brain heart infusion broth and 1 ml of the culture was centrifuged to collect the pellet. The pellet was suspended in 100 μl of distilled water into eppendorf tubes. The cellular suspension was brought to a boil for 10 min, and immediately was centrifuged at 14,000 RPM for 5 min. The supernatant was directly used for the PCR assay. The coagulase (Coa) and Protein A (Spa X region) gene specific primers (Table –1), synthesized by Merck (India) were used. PCR conditions used for amplification of Coa gene were: 94°C for 1 min, followed by 30 cycles of 94°C for 1 min, 58°C for 45 sec and 72°C for 1 min, followed by a final extension of 72°C for 10 min. and for amplification of Spa gene were: 94°C for 1 min, followed by 30 cycles of 94°C for 1 min, 60°C for 1 min and 72°C for 1 min, followed by a final extension of 72°C for 10 min. The amplified PCR products were detected by electrophoresis in 1.5% agarose gel prepared in 0.5X TBE buffer as per the method of Sambrook et al. (1989) and analyzed using Gel documentation system (Uvi Tech).
Antimicrobial sensitivity of the isolates was tested for Tetracycline, Offloxacin, Cephotaxime, Ciprofloxacin, Ampicilline, Lincomycin and Cephalexin using the modified disc diffusion method of Bauer et al. (1966).
RESULTS AND DISCUSSION
In present study, out of 214 quarter milk samples collected from lactating cows, 110 (51.40%) samples were found positive in California Mastitis Test (CMT). Of these 18 (8.41%), 24 (11.21%), 52 (24.30%) and 16 (7.48%) samples exhibited trace, weak positive, distinct and strong positive CMT reactions respectively. CMT has been recognized as a highly sensitive test to detect bovine SCM (Dangore et al., 2000; Madut et al., 2009).
Staph. aureus was isolated from all 68 samples that showed distinct (52) and strong positive reaction (16) in CMT. The Staph. aureus has been reported as the major pathogen involved in bovine mastitis (Singh and Baxi, 1982; Hameed et al., 2006; Sudhakar et al., 2009; Ranjan et al., 2011) and its association with subclinical mastitis as a major pathogen has been reported from various parts of the country (Ranjan et al., 2011). Out of 68 isolates tested, 65 (95.59 %) isolates showed catalase positive reaction. In Coagulase test all the isolates showed Coagulase activity. Of these, 54 (79.41%) and 56 (82.35 %) samples were found positive in slide and tube Coagulase test respectively. Further, 12 (17.65%) samples were positive for tube Coagulase test only and 14 (20.59) samples were positive in slide test, while 42 (61.76 %) samples exhibited positive reaction both in slide as well as tube Coagulase tests. In sugar fermentation reaction the mannitol was fermented by all the 68 isolates with production of acid and gas, while glucose was fermented with production of acid and gas by 59 (86.74 %) and 48 (70.58 %) isolates and lactose was fermented by 53 (77.94 %) and 52 (76.47 %) isolates respectively. On blood agar, 48 (70.58 %) isolates showed β– haemolysis and 59 (86.76 %) isolates were positive for nitrate reduction tests.
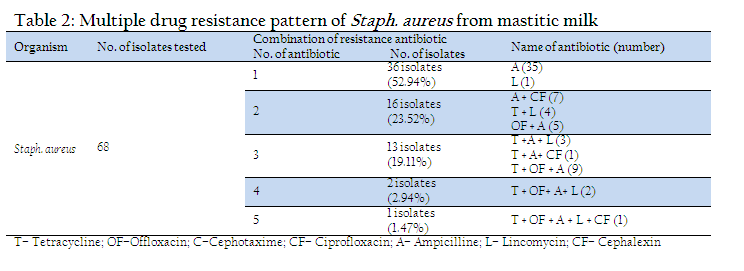
The coagulase production is important phenonotypic determinant of Staph. aureus association with virulence (Loebe, 1903; Jeljaszewicz et al.,1983). Coagulase positive Staphylococci were reported to be mostly positive to mannitol fermentation (Higgins and Chartier, 1984, Erasmus, 1985). β haemolysis is another trait used extensively for characterization of pathogenic Staph aureus. (Arshad et al., 2006, Franco et al., 2008). Bovine mastitis is the single most common ailment for the use of antibiotics in dairy cattle (Kaneene and Miller, 1992; Moore and Heider, 1984). Out of 68 isolates tested highest number of isolates (63, 92.64 % were resistant against ampicillin followed by 20 (29.41 %) for tetracycline and 17 (25%) for offloxacin. Eleven (16.7%) isolates showed resistant for lincomycin and 9 (13.23%) were found to be resistant for ciprofloxacin. All the isolates were found to be sensitive to cefotaxime. Out of 68 isolates tested, 36 isolates were resistant to single antibiotic (Table–2). Of these, 36 (52.94 %) isolates were resistant to ampicillin and one for lincomycin. Remaining 32 isolates exhibited multiple drug resistance involving two and more antibiotics. Of these, 16 (23.52 %) isolates were resistant to two antibiotics, 13 (19.11 %) to three antibiotics, 2 (2.94 %) to four antibiotics and one (1.47 %) isolate exhibited resistance to five antibiotics. The most common group of multiple drug resistance was observed to be tetracycline + offloxacin + ampicillin recorded in 9 isolates, followed by ampicillin + ciprofloxacin (7 isolates), ampicillin + offloxacin (5 isolates). One isolate showed multiple resistance for 5 antibiotics viz. tetracycline + offloxacin + ampicillin + lincomycin + Ciprofloxacin. The resistance of Staph. aureus to antimicrobial agents has been extensively documented and it contributed significantly to the treatment failure (Sumathi et al., 2008; Sudhakar et al., 2009; Kumar et al., 2010). The high resistance of Staph. aureus to ampicillin may be attributed to the production of betalactamase, an enzyme that inactivates penicillin and closely related antibiotics (Abera et al., 2010). It is believed that around 50% of mastitis causing Staph. aureus strains produce betalactamase (Green and Bradely, 2004).
In PCR, the Coa gene could be amplified in all the 68 isolates with a uniforn amplicon size of 710 bp. The presence of Coa gene in all the Staph aureus isolates is due to the fact that all the isolates included in the study, are caogulase producers as evident in biochemical tests. In the last few years, the Coa gene analysis has been extensively used for typing Staph. aureus isolates identified from bovine mastitis cases (Aarestrup et al., 1995; Lange et al., 1999; Schlegelova´et al., 2003). In present study, 22 (32.35%) isolates yielded a uniform Spa (X region) gene product of 320bp. Staphylococcal protein A (Spa) is an important virulence factor of Staph. aureus. The Spa gene of Staph. aureus encodes protein A and is used for typing of Staph. aureus (Harmsen et al., 2003). The isolates showing Spa gene were also positive for coagulase, catalase and nitrate tests and all fermented glucose, lactose and mannitol sugars on biochemical characterization. Single–locus DNA sequencing of repeat regions of the Coa and the Spa gene could be used for reliable and accurate typing of Staph. aureus strains (Frenay et al., 1996; Shopsin et al., 2000; Tang et al., 20002).
REFERENCES
Aarestrup FM, Dangler CA, and Sordillo LM. (1995). Prevalence of coagulase gene polymorphism in Staph. aureus isolates causing bovine mastitis. Can. J. Vet. Res. 59:124-128.
PMid:7648524 PMCid:PMC1263749
Abera M, Demie1 B, Aragaw K, Regassa F and Regassa A. (2010). Isolation and identification of Staph. aureus from bovine mastitic milk and their drug resistance patterns in Adama town, Ethiopia. J. Vet. Med. Anim. Hlth. 2: 29-34.
Arshad M, Muhammad G, Siddique M, Ashraf M Khan HA. (2006). Staphylococcal mastitis in bovines and some properties of staphylococcal isolates. Pakistan Vet. J. 26: 20-22.
Bauer AW, Kirby WMM, Sherris JC and Turck M. (1966). Antibiotic sensitivity testing by a standardized single disc diffusion method. Am. J. Clin. Patho. 45: 493-496.
PMid:5325707
Dangore AD, Bhalerao DP, Jagadish S, Keskar DV and Sharma LK. (2000). Evaluation of some byre side tests in bovine sub clinical mastitis. Indian Vet. J. 77: 380-381.
Devries LA and Oeding, P. (1976). Characteristics of Staph. aureus strains isolated from different animal species. Rev. Vet. Sci. 21: 284-294.
Erasmus JA. (1985). Some features of coagulase positive staphylococci from bovine milk. I. Carbohydrate metabolism, comparison of conventional techniques and the API 50 CH system. Onderrstepoort. J. Vet. Res. 52: 25-29.
PMid:3925403
Franco JC, Gonzalez L, Gomez SC, Carrillo JM and Ramirez JJ. (2008). eGnosis, 06: 1666-577745.
Frenay HM, Bunschoten AE, Schouls LM, van Leeuwen WJ, Vandenbroucke-Grauls JM, and Mooi, FR. (1996). Molecular typing of methicillin-resistant Staph. aureus on the basis of protein A gene polymorphism. European J. Clin. Microbiol. and Infect. Dis. 15: 60-64.
http://dx.doi.org/10.1007/BF01586186
PMid:8641305
Frenay HM, Theelen JP, Schouls LM, Vandenbroucke-Grauls JM, Verhoef J, Van Leeuwen WJ and Mooi FR (1994). Discrimination ofepidemic and nonepidemic methicillin-resistant Staph. aureus strains on the basis of protein A gene polymorphism. J. Clin. Microbiol. 32:846–847.
PMid:8195406 PMCid:PMC263139
Gao J and Stewart GC (2004). Regulatory Elements of the Staph. aureus Protein A (Spa) Promoter. J. Bacterial. 86: 3738–3748.
http://dx.doi.org/10.1128/JB.186.12.3738-3748.2004
PMid:15175287 PMCid:PMC419937
Gianneechini R, Concha C, Rivero R, Delucci I and Morenolpez J (2002). Occurrence of clinical and subclinical mastitis in dairy herds in West Littoral region in Urugway. Acta Vet. Scand. 43: 221-230.
http://dx.doi.org/10.1186/1751-0147-43-221
http://dx.doi.org/10.1186/1751-0147-43-31
PMid:12831175
Green M and Bradely A (2004). Clinical forum Staph. aureus mastitis in cattle UK Vet. 9: 4.
http://dx.doi.org/10.2202/1540-8884.1053
Hameed KGA and Sender G (2006). Public health hazard due to mastitis in dairy cows. Anim. Sci. Pap. & Rep. 25: 73-85
Harmsen D, Claus H, Witte W, Rothga¨nger J, Claus, H, Turnwald D and Voge U (2003). Typing of methicillin-resistant Staph. aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442–5448.
http://dx.doi.org/10.1128/JCM.41.12.5442-5448.2003
PMid:14662923 PMCid:PMC309029
Higgins R and Chartier P (1984). Identification of coagulase positive staphylococci of animal origin. Medicine Veterinarie du Quebec, 14: 61-65.
Himabindu M, Muthamilselvan DS, Bishi DK and Verma RS (2009). Molecular Analysis of Coagulase Gene Polymorphism in Clinical Isolates of Methicilin Resistant Staphylococcus aureus by Restriction Fragment Length Polymorphism Based Genotyping. American J. Inf. Dis. 5: 170-176.
http://dx.doi.org/10.3844/ajidsp.2009.170.176
http://dx.doi.org/10.3844/ajidsp.2009.163.169
Jeljaszewicz J, Switalski LM, Adlam C (1983). Staphylo coagulase and clumping factor. In: Easmon CSF, Adlam C, eds. Staphylococci and Staphylococcal Infections. Academic Press, London. 2. 525-558.
Kaneene JB and Miller R (1992). Description and evaluation of the influence of Veterinary presence on the use of antibiotics and sulfonamides in dairy herds. J. Am. Vet. Med. Assoc. 201: 68-76.
PMid:1644649
Kumar R, Yadav BR and Singh RS (2010). Genetic Determinants of antibiotic resistance in Staph. aureus isolates from milk of mastitic crossbred cattle. Curr. Microbiol. 60: 379–386
http://dx.doi.org/10.1007/s00284-009-9553-1
PMid:19957184
Lange C, Cardoso M, Senczek D and Schewarz S (1999). Molecular subtyping of Staphylococcus aureus isolates from cases of bovine mastitis in Brazil. Vet. Microbiol. 67: 127-141
http://dx.doi.org/10.1016/S0378-1135(99)00031-0
Loebe L (1903).The influence of certain bacteria on the coagulation of the blood. J Med Res 10: 407-419.
Madut NA, Godir AEA and El Jalil IM (2009). Host determinants of bovine mastitis in semi-intensive production system of harfoum State, Sudan. Journal of Cell and Animal Biology, 3: 71-77.
Moore GA and Heider LE (1984). Treatment of mastitis. Vet. Clin. North Am. Large Anim. Prac. 6: 323-331.
PMid:6474755
National Mastitis Council (NMC) (1990). Microbiology procedures for the diagnosis of udder infection 3rd ed. Arlington. V.A. National Mastitis Council Inc.
Quinn PJ, Carter ME, Markey B and Carter GR (1994). Clinical veterinary microbiology. Ist Edn. London, U.K., Mosby Publication pp 118-137.
Ranjan R, Gupta MK and Singh KK (2011). Study of bovine mastitis in different climatic conditions in Jharkhand, India. Vet. World 4: 205-208.
http://dx.doi.org/10.5455/vetworld.2011.205-208
Riffon R, Saysith K and Khalil H (2001). Development of a rapid and sensitive test for identification of major pathogens in bovine mastitis by PCR. J. Clin Microbiol. 39: 2584-2589.
http://dx.doi.org/10.1128/JCM.39.7.2584-2589.2001
PMid:11427573 PMCid:PMC88189
Sambrook J, Fritsch EF and Maniatis T (1989). Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
Saravanan P, Nagarajan B, Ramprabhu R, Vashu K and Dhanapalan P (2000). A study on etiology, incidence and physical character of milk in sub-clinical mastitis. Ind. J. Vet. Med. 20: 76.
Schlegelová J, Dendis M, Benedík J, Babák V, Rysánek D. (2003). Staph. aureus isolates from dairy cows and humans on a farm differ in coagulase genotype. Vet Microbiol. 92:327–334.
http://dx.doi.org/10.1016/S0378-1135(02)00409-1
Shopsin B, Mathema B, Martinez J, Ha E, Campo ML, Fierman A, Krasinski K, Kornblum J, Alcabes P, Waddington M, Riehman M and Kreiswirth BN (2000). Prevalence of methicillin-resistant and methicillin-susceptible Staph. aureus in the community. J. Infect. Dis. 182:359-362.
http://dx.doi.org/10.1086/315695
PMid:10882625
Singh KB and Baxi KK (1980). Studies on the incidence and diagnosis of subclinical mastitis in milch animals. Indian Vet. J. 57: 723 – 729.
Strommenger B, Kehrenberg C; Kettlitz C, Cuny C, Versppohl J, Witte W and Schwarz S (2006). Molecular characterization of methicilin resistant Staph. aureus strain from pet animals and their relationship to human isolates. J. Antimicrobial Chemo. 57: 461-465.
http://dx.doi.org/10.1093/jac/dki471
PMid:16387748
Su C, Kanevsky I, Jayarao BM, and Sordillo LM. (1999). Phylogenetic relationships of Staph. aureus from bovine mastitis based on coagulase gene polymorphism. Vet Microbiol. 7:53-58.
Sudhakar P, Awandkar N and Khode V (2009). Prevalence and current antibiogram trend of mastitic agents in Udgir and its vicinity, Maharastra state, India. Int. J. of Dairy Sci. 4: 117-122.
http://dx.doi.org/10.3923/ijds.2009.117.122
Sumathi BR, Veeregowda BM and Amitha RG (2008). Prevalence and antibiogram profile of bacterial Isolates from clinical bovine mastitis: Vet. World, 8:237- 238.
Tang YW, Waddington MG, Smith DH, Manahan JM, Kohner PC, Highsmith LM, Li H, Cockerill FR Thompson RL, Montgomery SO and Persing DH (2000). Comparison of protein A gene sequencing with pulsed-field gel electrophoresis and epidemiologic data for molecular typing of methicillin-resistant Staph. aureus. J. Clin. Microbiol. 38: 1347–1351.
PMid:10747105 PMCid:PMC86443
Van Leeuwen WJ, Belkum V, Verkooyen AR, Saçilik SC, Cokmus C and Verbrugh H (1997). Dissemination of a single clone of methicillin-resistant Staph. aureus among Turkish hospitals. J. of clin. Mcrobiol. 35: 978–981.
Vandensesch F, Naimi T, and Enright M.C. (1994). Community-acquired methicillin resistant Staph. aureus carrying panton Valentine Leukocidin genes: worldwide emergence. Emerg Infect Dis., 9: 978-984.
http://dx.doi.org/10.3201/eid0908.030089
PMid:12967497 PMCid:PMC3020611
Wani SA, Bhatt MA, Munshi ZH, Wuereshi S and Buchh AS (2003). Isolation and in-vitro sensitivity pattern of pathogenic Eschericha coli from diarrhoeic faecal samples of lambs and calves. Ind. J. Animal Sci., 73: 168-170.