Journal of Animal Health and Production
Research Article
Journal of Animal Health and Production 2 (1): 1 – 4Prevalence and Antimicrobial Resistance Profile of Escherichia Coli and Salmonella Isolated from Diarrheic Calves
Abu Raihan Muhammad Ismail Hossain Ansari1, Muhammad Mostafizer Rahman2, Muhammad Zohorul Islam3, Bhajan Chandra Das4, Ahasan Habib5, S.M. Shariful Hoque Belal6, Kamrul Islam3*,
- Veterinary Surgeon, District Veterinary Hospital, Nilphamari;
- Department of Microbiology, Faculty of Veterinary and Animal Science, Haji Mohammad Danesh Science and Technology University, Dinajpur;
- Department of Microbiology, Faculty of Veterinary Medicine, Chittagong Veterinary and Animal Sciences University, Chittagong;
- Department of Medicine and Surgery, Faculty of Veterinary Medicine, Chittagong Veterinary and Animal Sciences University, Chittagong;
- Department of Parasitology, Bangladesh Agricultural University, Mymensingh;
- Veterinary Surgeon, District Veterinary Hospital, Sirajgonj, Bangladesh;
*Corresponding author: kamruldvm13@gmail.com
ARTICLE CITATION:
Ansari ARMIH, Rahman MM, Islam MZ, Das BC, Habib A, Belal SMSH, Islam K (2014). Prevalence and antimicrobial resistance profile of Escherichia coli and salmonella isolated from diarrheic calves. J. Anim. Health Prod. 2 (1): 12 – 15.
Received: 2014–03–08, Revised: 2014–04–21, Accepted: 2014–04–23
The electronic version of this article is the complete one and can be found online at
(
http://dx.doi.org/10.14737/journal.jahp/2014/2.1.12.15
)
which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
ABSTRACT
Neonatal calf diarrhea (NCD) is a common disease affecting the newborn calf and the most critical period is in the first few days following birth of the calf which is also known as calf scours. Keeping animals in close confinement where the opportunity for transmission of causative agents of NCD. The diarrhea and other clinical signs seen with the disease are caused by the interaction of any of several possible infectious causes. This study was carried out to isolate, identify and detect the antimicrobial resistant profile of E. coli and Salmonella from diarrheic calves. A total of one hundred and twenty five fecal specimens were collected directly from the rectum of diarrheic calves. Of the samples collected 35 (25%) and 11 (8.8%) was found positive for E. coli and Salmonella respectively. Antimicrobial resistance of these two isolate was found against Amoxycillin and Tetracycline whereas a high sensitivity was found towards Ciprofloxacin, Levofloxacillin, Azithromycin and Cefotaxime. Serotyping was done by using specific antisera to identify variants of the somatic (O) and flagellar (H) antigens. Cultural and biochemical features also reveal the presence of pathogens in the diarrheic calves.
INTRODUCTION
For the newborn calf one of the most critical periods is the first week of life and is generally associated with a mortality rate of 10%. Diarrhea is one of the major cause of mortality in newborn calves, the incidence of diarrhea in calves less than one month ranges between 15 to 20%, signifying that the greatest risk occurs during the first two weeks of life (Vandeputte et al., 2010). Calf diarrhea is a major cause of economic loss with high morbidity and mortality in the cattle industry worldwide (Kelling et al., 2002; Uhde et al., 2008; Bartels et al., 2010). In Bangladesh, calf diarrhea remains the most often reported clinical problem in calf management and rearing system (Debnath et al., 1990). Historically, calf diarrhea has been commonly attributed to bovine rotavirus group A (BRV–A), bovine corona virus (BCoV), bovine viral diarrhea virus (BVDV), Salmonella spp. (Salmonella), Escherichia coli (E. coli), and Clostridium perfringens (C. perfringens) type C and Cryptosporidium parvum (C. parvum) (Safi and Smith, 1985; Reynolds et al., 1986; Snodgrass et al., 1986; Acha et al., 2004;). However to recover this significant economic loss, heavy amounts of antimicrobials are used in calves feed as a preventive and curative purposes worldwide (Dheilly et al., 2011). The inevitable selection of antimicrobial compound that results resistance in calf pathogens and commensals may emerge and become a worldwide public health problem through impact on food safety which led to failure of prevention and treatment. Antimicrobial–resistant bacteria carried by animals can enter the human food chain through the consumption of meat or other animal products, through farm runoff water, and by other pathways (Donnelly, 1999; Tiwari et al., 2013). The study was conducted with objectives: to isolate and identify the bacteria associated with calf diarrhea; to characterize the bacteria by different cultural, biochemical and serological tests; and to study the antibacterial sensitivity of the identified field isolates.
MATERIALS AND METHODS
Collection of Samples
A total of one hundred and twenty–five fresh fecal samples were collected from calves suffering from diarrhea and enteritis. The samples were collected from the selected calves and sent to the laboratory for microbiological investigations.
Isolation of Bacteria
Firstly fecal samples were inoculated into nutrient broth (NB) and incubated at 37°C for 24 hours and then the growth were inoculated into nutrient agar (NA) and incubated at 37°C for 24 hours. The cultivated organisms from NA agar were inoculated directly into MacConkey agar and incubated at 37°C for 24 hours. Lactose fermenting pink (bright red) colony from the MacConkey agar was sub–cultured into selective media (EMB agar) and incubated at 37ºC for 24 hours. The non lactose fermenting colorless colony from the MacConkey agar was sub–cultured on SS agar media and on Brilliant green agar (BGA) media used as a selective media for pathogenic Salmonella and incubated at 37ºC for 24 hours. .
Microscopic Study by Staining Method
Grams staining method was done to study morphology and staining characters. A Suspected colony from EMB agar and SS agar were stained as described by Singh and Prekash (2008).
Identification of bacterial isolates by using specific biochemical tests
Various biochemical tests were performed for species identification. For this study isolated organisms with supporting growth characteristic of E. coli on EMB and Salmonella on SS and BGA were subjected to various biochemical tests named carbohydrate fermentation tests, TSI agar slant reaction, MR–VP, MIU, Indole reaction and citrate utilization test were carried out for identification of suspected Salmonella. All the isolates from different sources were tested for the detection of E. coli and Salmonella.
Serotyping by Slide Agglutination Test
The polyvalent agglutinating antiserum poly “O” and poly “H” against Salmonella manufactured by S and A Reagents Lab, Bangkok, Thailand, was used for the serotyping of the isolated Salmonella . The macroscopic slide agglutination tests were performed. The cultures to be tested were first checked with salmonella poly “O” polyvalent antiserum. A single isolated colony from BG agar was dissolved in physiological saline solution. One drop of thick bacterial suspension was placed on glass slide and a drop of polyvalent antiserum was added. The slide was gently rotated to mix the contents thoroughly. Those cultures which agglutinated within one to two minutes were selected as positive for Salmonella and subjected to agglutination test with Salmonella agglutinating antiserum (poly “H”).
Antibacterial Sensitivity Pattern of the Isolated Salmonella and E. coli
The overnight nutrient broth cultured Salmonella isolates were poured on SS agar and spread uniformly with the help of sterile glass spreader. Antibacterial discs were applied aseptically to the surface of the plate at an appropriate distance with the help of sterile forceps and incubated at 37ºC for 24 hours, aerobically. Antibiotic sensitivity pattern of isolated E. coli and Salmonella were performed against 14 commonly used antibiotics belonging to different groups (Bauer et al., 1966).
RRESULT
Following Gram’s staining technique, the smear revealed gram negative rods of different shape and size arranged in single, paired or in short chain manner indicating possibility of E. coli while another smear showed small, uniform rod shaped gram negative organisms arranged singly and sometimes in pairs indicating probability of Salmonella . On nutrient agar isolated E. coli produced smooth, circular and white to grayish white colony with peculiar fetid odor and Salmonella produced circular, smooth, opaque and translucent colonies. E. coli produced bright pink or red colonies over MacConkey agar while the Salmonella showed colorless, smooth, pale, transparent colonies. On EMB agar the fecal isolates of E. coli produced raised, large, smooth and sticky colony with yellow green metallic sheen. E. coli produced pinkish colony and the isolated Salmonella exhibited opaque, translucent and colorless colonies on SS agar. On BGA E. coli produced yellowish green color and the isolated Salmonella produced pale pink color colonies against a pinkish background which was earlier green in color before growth.
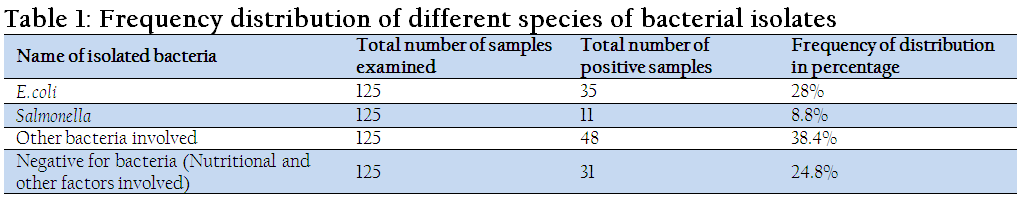
The results of frequency distribution of bacterial isolates were presented in Table 1. A total of 125 fecal samples were examined for the isolation of bacteria, of which 35 (28%) samples were positive for E. coli, 11 (8.8%) samples were positive for Salmonella and 31 (24.8%) samples were negative for any bacteria.
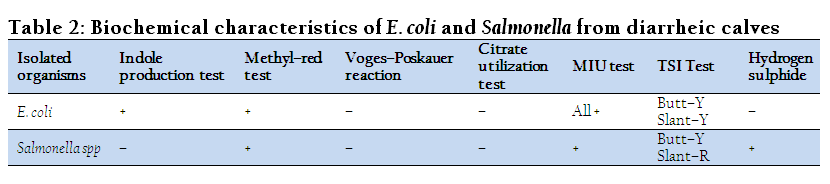
Bacteria isolated from feces of diarrheic calves were subjected to various physio–chemical tests to determine their biochemical characters and degree of variation in their reactivity pattern. The results of these tests are presented in Table 2. The isolated salmonella gave positive agglutination test with Salmonella agglutinating antiserum poly “O” and “H”.
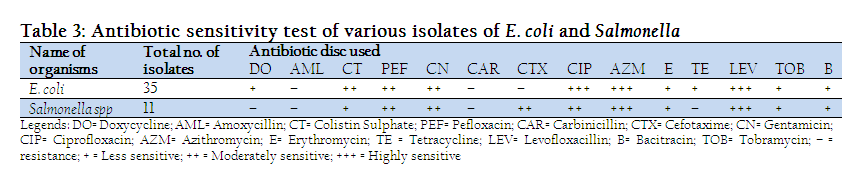
From the antibiogram study, it was revealed that among the isolated E. coli organism from diarrheic samples of calves 100% were highly sensitive to Azithromycin, Ciprofloxacin and Levofloxacillin. Cent percent bacteria were moderately sensitive to Colistin sulphate & Pefloxacin, 80% to Gentamicin, 20% to Cefotaxime. On the other hand all the tested bacteria were less sensitive to Tobramycin, 80% were less sensitive to Doxycycline, Bacitracin & Erythromycin; 20% were less sensitive to Gentamicin, Cefotaxime, Carbinicillin. 100% were resistant to Amoxycillin and Tetracycline, whereas 80% resistant to Carbinicillin, 60% resistant to Cefotaxime and 20% were resistant to Bacitracin & Erythromycin. Among the isolates of Salmonella spp. 100% were highly sensitive to Levofloxacillin, 75 to Ciprofloxacin, 50% to Azithromycin. 100% were moderately sensitive to Pefloxacin and Cefotaxim, 75% were to Gentamicin, 50% were to Azithromycin and 25% were moderately sensitive to Ciprofloxacin, Colistin Sulphate and Erythromycin. 75% were less sensitive Bacirtracin and Colistin Sulphate, 50% less sensitive to Tobramycin but 25% were less sensitive to Doxycycline, Gentamicin, Carbinicillin and Erythromycin. Besides those, 100% were resistant to Amoxycillin & Tetracycline, 75% to Carbinicillin and Doxycycline, 50% to Erythromycin and Tobramycin, 25% were to Bacitracin (Table 3).
DISCUSSION
In the present investigation, a total of one hundred and twenty–five fresh fecal samples were collected from calves suffering from diarrhea and enteritis. Of which 35 samples were found positive for E. coli gives a positive reaction to lactose fermentation on MacConkey agar plate, metallic sheen colonies on EMB plates and yellowish green colonies on BGA, 11 samples were found positive for Salmonella, producing negative reaction to lactose fermentation on MacConkey agar plate. Opaque, translucent and colorless colonies on SS agar, pale pink color colonies against a pinkish background over BGA and deep blue color on green color Simmons citrate agar. Similar cultural characteristics were also corroborated by (Abdullah et al., 2013). Gram staining were performed for all the isolates and revealed Gram negative, non–acid fast, uniformly stained, non–spore forming bacilli. These findings were identical with the earlier studies performed by other workers (Merchant and Packer, 1967). Serotyping of salmonella based on the agglutination of bacteria with specific sera to identify variants of the somatic (O) and flagellar (H) antigens is supported by earlier work of (Wattiau et al., 2011).
The frequency distributions of different species of bacterial isolates in different fecal samples were found variable. The results of the present study indicated that two different types of bacteria were present in the fecal samples collected from diarrheic calves. Of the samples collected 35 (28%) and 11 (8.8%) were found positive for E. coli and Salmonella respectively. The observations about prevalence of these bacterial organisms were supported by a recent study (Abdullah et al., 2013), who out of 114 fecal samples, 44 (38.6%) samples were found positive for E. coli and 25 (21.9%) samples for Salmonella spp.
The different isolates of E. coli and Salmonella showed identical results in different biochemical tests i.e., TSI, MIU, Indole, MR–VP and citrate utilization tests. This type of similarity may be due to presence of some common genetic materials that could manifest the similar types of biochemical strategy (Abdullah et al., 2013).
The in vitro antibiotic sensitivity assay of both bacterial isolates to different antibiotics was carried out. A slight variation was noticed in the results of the sensitivity of isolates against 14 different antibiotics used. The isolated Salmonella and E.coli bacteria were highly sensitive to levofloxacillin, ciprofloxacin, azithromycin, cefotaxime; moderately sensitive to gentamicin, azithromycin, pefloxacin, cefotaxime, erythromycin, carbinicillin and to ciprofloxacin. They were less sensitive to tobramycin, bacitracin, erythromycin, doxycycline, tetracycline, carbinicillin, cefotaxime, while resistant to amoxycillin, tetracycline, bacitracin, tobramycin, doxycycline, carbinicillin, erythromycin and cefotaxime. The antibacterial resistance observed in the isolated Salmonellae and E. coli might be due to indiscriminate use of those antibacterial agents in the study areas or rapid chromosomal mutation and the presence of specific plasmid DNA. The results of study will provide guidelines to the veterinarian to select the appropriate antibiotics to reduce the economic losses by selecting the sensitive antibiotics. This finding correlate the results of some previous studies stated that calf isolates were highly sensitive to ciprofloxacin, levofloxacin and resistant to ampicillin, erythromycin, gentamicin and amoxicillin (Guerra et al., 2006; Ahmed et al., 2009).
The results of isolation, identification, biochemical test, frequency distribution, and antibiotic sensitivity of the bacteria isolated from calf diarrhea in the present study indicates that the microbial factors might play an important role for the development of calf diarrhea and alternative treatment approaches should be looked for (Dhama et al., 2013; Mahima et al., 2013).
CONCLUSION
Prevalence of E. coli was higher than Salmonella in diarrheic calves. The antimicrobial resistance profile was varied but Ciprofloxacin, Levofloxacillin, Azithromycin and Cefotaxime showed more sensitivity compared to other drugs.
CONFLICT OF INTEREST
Authors declare that they have no competing interests.
REFERENCES
Abdullah M, Akter MR, Lutful Kabir SM, Abu Sayed Khan M, Saleh Ibne S, Aziz MA (2013). Characterization of Bacterial Pathogens Isolated from Calf Diarrhoea in Panchagarh District of Bangladesh. J. Agric. Food. Tech. 3(6): 8–13.
Acha SJ, Kuhn I, Jonsson P, Mbazima G, Katouli M, Mollby R (2004). Studies on calf diarrhea in Mozambique: prevalence of bacterial pathogens. Acta Vet. Scand. 45: 27–36.
http://dx.doi.org/10.1186/1751-0147-45-27
PMid:15535084 PMCid:PMC1821001
Ahmed AM, Younis EE, Osman SA, Ishida Y, El–Khodery SA, Shimamoto T (2009). Genetic analysis of antimicrobial resistance in Escherichia coli isolated from diarrheic neonatal calves. Vet. Microbiol. 136(3–4):397–402.
http://dx.doi.org/10.1016/j.vetmic.2008.11.021
PMid:19128900
Bartels CJ, Holzhauer M, Jorritsma R, Swart WA, Lam TJ (2010). Prevalence, prediction and risk factors of enteropathogens in normal and non–normal faces of young Dutch dairy calves. Prev. Vet. Med. 93: 162–169.
http://dx.doi.org/10.1016/j.prevetmed.2009.09.020
PMid:19819574
Bauer AW, Kirby WM, Sherris JC, Turck M (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45(4):493–6. PMid:5325707
Debnath NC, Sil BK, Selim SA, Prodhan MA, Howlader MR (1990). A retrospective study of calf mortality and morbidity in small holder traditional farming in Bangladesh. Prev. Vet. Med. 9: 1–7.
http://dx.doi.org/10.1016/0167-5877(90)90037-I
Dhama K, Chakraborty S, Mahima, Wani MY, Verma AK, Deb R, Tiwari R, Kapoor S (2013). Novel and emerging therapies safeguarding health of humans and their companion animals: a review. Pak. J. Biol. Sci. 16(3): 101–111.
http://dx.doi.org/10.3923/pjbs.2013.101.111
PMid:24171271
Dheilly A, Bouder A, Devendec LL, Hellard G, Kempf I (2011). Clinical and microbial efficacy of antimicrobial treatments of experimental avian colibacillosis. Vet. Microbiol. 149:422–429.
http://dx.doi.org/10.1016/j.vetmic.2010.11.033
PMid:21185134
Donnelly JP (1999). Commentary on the MAFF technical report: a review of antimicrobial resistance in the food chain. Int. J. Antimicrob. Agents, 12:63–65.
http://dx.doi.org/10.1016/S0924-8579(99)00048-5
Guerra B, Junker E, Schroeter A, Helmuth R, Guth BG, Beutin L, Chemother AJ (2006). Phenotypic and genotypic characterization of antimicrobial resistance in Escherichia coli O111 isolates. J. Antimicrob. Chemother. 57(6):1210–4.
http://dx.doi.org/10.1093/jac/dkl127
PMid:16603644
Kelling CL, Steffen DJ, Cooper VL, Higuchi DS, Eskridge KM (2002). Effect of infection with bovine viral diarrhea virus alone, bovine rotavirus alone, or concurrent infection with both on enteric disease in gnotobiotic neonatal calves. Ame. J. Vet. Res. 63:1179–1186.
http://dx.doi.org/10.2460/ajvr.2002.63.1179
PMid:12171174
Mahima Ingle, AM, Verma, AK, Tiwari, R, Karthik K, Chakraborty S, Deb R, Rajagunalan S, Rathore R, Dhama K (2013). Immunomodulators in day to say life: a review. Pak. J. Biol. Sci. 16(17): 826–843.
http://dx.doi.org/10.3923/pjbs.2013.826.843
PMid:24498836
Merchant IA, Packer RA (1967). Veterinary Bacteriology and Virology. Seventh edi. The Iowa University Press, Ames, Iowa, USA, pp. 286–306.
Reynolds DJ, Morgan JH, Chanter N, Jones PW, Bridger JC, Debney TG, Bunch KJ (1986). Microbiology of calf diarrhea in southern Britain. Vet. Rec. 119: 34–39.
http://dx.doi.org/10.1136/vr.119.2.34
PMid:3750767
Singh P, Prakash A (2008). Isolation of Escherichia coli, Staphylococcus aureus and Listeria monocytogenes from milk products sold under market condition at Agra region. Acta agri. Slov. 92 (1): 83–88.
Snodgrass DR, Terzolo HR, Sherwood D, Campbell I, Menzies JD, Synge BA (1986). Aetiology of diarrhea in young calves. Vet. Rec. 119: 31–34.
http://dx.doi.org/10.1136/vr.119.2.31
PMid:3750766
Tiwari R, Chakraborty S, Dhama K, Rajagunalan S, Singh SV (2013). Antibiotic resistance – an emerging health problem: causes, worries, challenges and solutions – a review. Int. J. Curr. Res. 5(07): 1880–1892.
Uhde FL, Kaufmann T, Sager H, Albini S, Zanoni R, Schelling E, Meylan M (2008). Prevalence of four enteropathogens in the faeces of young diarrhoeic dairy calves in Switzerland. Vet. Rec. 163: 362–366.
http://dx.doi.org/10.1136/vr.163.12.362
PMid:18806281
Vandeputte S, Detilleux J, Care S, Bradfer B, Guyot H, Rollin F (2010). Evaluation of a Bovine Concentrated Lactoserum for Preventing Neonatal Diarrhea in Belgian Blue Calves. The Open Vet. Sci. J. 4: 36–40.
Wattiau P, Boland C, Bertrand S (2011). Methodologies for Salmonella enterica subsp. enterica Subtyping: Gold Standards and Alternatives. App. and Env. Microbiol. pp. 7877–7885.
PMid:21856826 PMCid:PMC3209009