Journal of Animal Health and Production
Short Communication
Journal of Animal Health and Production. 2 (1): 8 – 11Nucleotide Variationin Cation Channel of Sperm 1 Gene in Vrindavani Cattle
Thangavel Geetha, Subodh Kumar*
*Corresponding author: subkum@gmail.com
ARTICLE CITATION:
Geetha T and Kumar S (2014). Nucleotide variationin cation channel of sperm 1 gene in vrindavani cattle. J Anim Health Prod. 2 (1): 8 – 11.
Received: 2014–01–31, Revised: 2014–02–22, Accepted: 2014–02–23
The electronic version of this article is the complete one and can be found online at
(
http://dx.doi.org/10.14737/journal.jahp/2014/2.1.8.11
)
which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited
ABSTRACT
The present study was carried out on Vrindavani cattle, to investigatethe nucleotide variations in exon 3 and 4 of Cation channel of Sperm 1 (CatSper1) gene by the PCR–RFLP technique and the nucleotide sequencing of respective amplicons. To detect restriction site polymorphism, PCR–RFLP was performedon genomic DNA isolated from blood of 100 randomly selected crossbred animals. The PCR–RFLP of 482 bp encompassing complete exon 3 and 4 with flanking introns, using PvuII and RsaIrestriction enzymes produced two (403 and 79 bp) and three bands (229, 152 and 103 bp), respectively. This fragment of CarSper1 gene exhibited absence of polymorphism with respect to restriction enzymes used and accordingly, the allelic frequency was found to be 1.0. The monomorphic pattern the amplicon of CatSper1 gene with respect to different enzymes indicated the conservednessof this gene. Ampliconswere subjected to DNA sequencing after which it was annotated and submitted to GenBank. In order to study the variations at nucleotide level, the sequences of studied amplicons were compared with similar sequences of Zebu cattle, buffalo and goat.
Indiahas been witnessing steady increase in crossbred cattle population,which is contributing significantly to the success of dairy industry. To sustain the increasing population of crossbreds, more doses of crossbred semen are required. Semencryo–preservation becomes a tool of importance in such a situation.The ultimate goal of semen preservation has been to obtain pregnancies after artificial insemination, which would be as effective as natural mating. The successful preservation of semen depends on several factor, which are involved in the process of cryopreservation and the term ‘freezability’ has been given to mean the ability of semen to survive after being frozen without suffering substantial damage. The damage to the semen expressed as sperm count, motilityand fertility but practically, it largely refers to post thaw motility (PTM) (Ravimuruganet al., 2007) which again depends on initial/pre–freeze motility and is the ultimate trait which qualifies the semen for artificial breeding. Crossbred bulls have high percentage of abnormal spermatozoa, lower level of sperm motility and viability, together causing decline in fertility rate (Dhanjuet al., 2006). Poor sperm motility and freezability of semen has also been reported in Vrindavani crossbred bulls (Ghosh et al., 2007). Researchers have reported more than 50% rejection rate in different crossbred bulls (Chacon et al., 1999; Tyagiet al., 2006) due to poor quality,especially the sperm motility. Many genes are known to control sperm motility.Off late, an ion channel gene was identified which had a significant bearing on sperm motility (Ren et al., 2001; Quill et al., 2001). Thiswas named as Cation channel of sperm (CatSper) and reported to have four subunits.CatSper family mutations resulted in male infertility (Darszonet al., 2006).CatSpers1–4 are expressed in testis and localized primarily to the principal piece of sperm tail (Ren et al., 2001; Quill et al., 2001; Lobleyet al., 2003; Jin et al., 2005). CatSper1 and 2 are required for the hyperactivation of sperm cell motility, which is essential for fertility(Qi et al., 2007). This study was therefore taken up, to ascertain nucleotide variation inCatSper1 gene inVrindavani crossbred cattle, so as to initiate a step in searching the promising DNA marker that could be developed to improve sperm motility of crossbred cattle by assisting in bull selection process.
A total of hundred and five randomly selected Vrindavani cattle(50–62.5 % exotic inheritance comprising of Holstien Frisian, Brown Swiss, Jersey; with Hariana as indigenous stock) developed at the Indian Veterinary Research Institute, Izatnagarwere included in the present investigation. A total of 15 ml of venous bloodwas collected from each crossbred animal in sterile 15 ml polypropylene centrifuge tube containing 0.5 ml of 2.7% EDTA as anticoagulant. DNA isolation was performedby phenol chloroform extraction method (Sambrook and Russel, 2001) and the precipitated DNA was dissolve in 200µl of TE buffer. The quality and concentration of genomic DNA was evaluated by spectrophotometer (PG Instruments, UK). Samples that showed an OD ratio (OD260/OD280) in the range of 1.7 to 1.9 were assessed to be of good quality.
Primerswere designed by the help of DNASTAR software to amplify CatSper 1 gene on the basis of already reportedsequence (in silico generated/ predicted)in Bostaurus cattle (NC_007330) available in the GenBank (www.ncbi.nlm.nih.gov).The fragment of CatSper 1 gene (482bp) comprising of complete exon 3 and 4 with flanking introns, was amplified by using a pair of self designedprimers (Forward: 5' CCT CAC CAC GGC GAA CAC CAC 3' and Reverse: 5' GGA CTA CAC CAG CAG GGG AGA GC 3'). The stock solution was prepared by diluting the same with DNase free water (Biogene, USA) in such a way that each has concentration of 300pmoles/μl. This was kept at 4oC for 2–3 days for allowing complete dissolution of primers. The working primer solution was further prepared by 10 fold dilution of stock primer solution in DNAse free water (Biogene, USA) so that each has a concentration of 30 pmoles/μl.A master mix was prepared for the required number of reactions by adding the reaction components in the following order, autoclaved distilled water, 10X assay buffer, dNTPs, MgCl2,forward and reverse primers,and finally Taq DNA polymerase enzyme. The 24µl of master mix was distributed into 0.2 ml of each PCR tubes duly labeled and marked. Finally, 1 µl of genomic DNA was added to the PCR tubes followed by gently mixing. The PCR tubes with the reaction mixture were put in a thermocycler (iCycler, Biorad, USA). The reaction mixture and PCR programme were followed as suggested by Sharma et al. (2009, 2010), to achieve the satisfactory level of amplification in a final volume of 25μl containing genomic DNA (60–100ng), 2.5μl of 10xPCR (1.5mM), 2.5 μl of dNTPs mix (0.2mM), 1.5 μl of MgCl2 (1.5mM), 1μl each forward and reverse primers and 0.2μl of Taq DNA polymerase (5 U/μl).Samples were amplified for 35 cycles with initial denaturation at 94oC for 3 min., cyclic denaturation 94oC for 1min., annealing at 62oC for 1min., cyclic extension 72oC for 1min., final extension 72oC for 10min.
Theamplicon was digested by PvuI and RsaIenzymes, which had the recognition frames as CAG↓CTG and GT↓AC respectively. The digested products were electrophoresed in 2.5% w/v agarose gel, stained with ethidium bromide, at 100 V for 5min and then 90V for 1 hr in 1x TBE buffer and visualized under UV light. The amplicon of CatSper1 gene was eluted, cloned in pGEMT vector and sequenced in both orientations. The obtained sequence on crossbred cattle was then aligned with similar sequences in other species viz. zebu cattle (Bosindicus), buffalo (Bubalusbubalis) and goat(Capra hircus) using MEGALIGN module of DNASTAR software (Lasergene, USA).
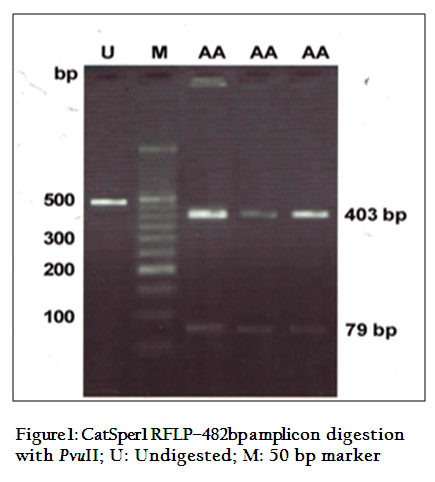
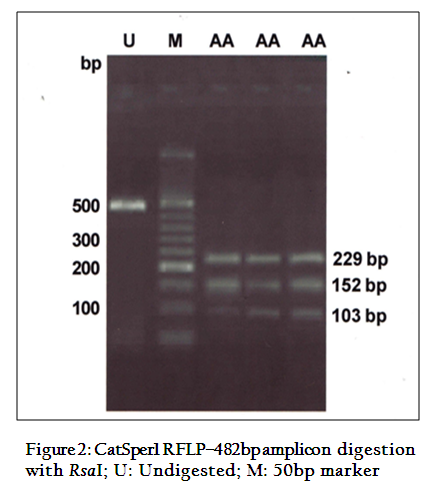
The digestion of 482 bpamplicon of encompassing exon 3 and 4, with PvuII produced two bands of 403 and 79bp (Figure 1). On digestion of same PCR products withRsaI gave three bands of 229, 152 and 103bp (Figure 2) respectively. This restriction pattern was due to the presence of a singlePvuII site (CAG↓CTG) at the 403rd position and two RsaI site (GT↓AC) at the 103rd and 255th position of amplicon. The DNA samples showed the same restriction pattern depicting the presence of same RE site in all animals. The results suggested that this amplicon was monomorphic, since all of them showed a single homozygous genotype.Monomorphic pattern has also been observed in exon 2 using EcoRI, HindIIIand exon 5 usingAluI and TaqI restriction enzymes of CatSper1 gene in Vrindavani cattle (Geethaet al., 2011, 2013). However, polymorphism was observed in this gene by using PCR–SSCP method by Modiet. al. (2011, 2014).Sivakumaret al. (2013a, 2013b) also observed various SNPs in CatSper1 and CatSper2 genes in vrindavani as well as tharparkar cattle.
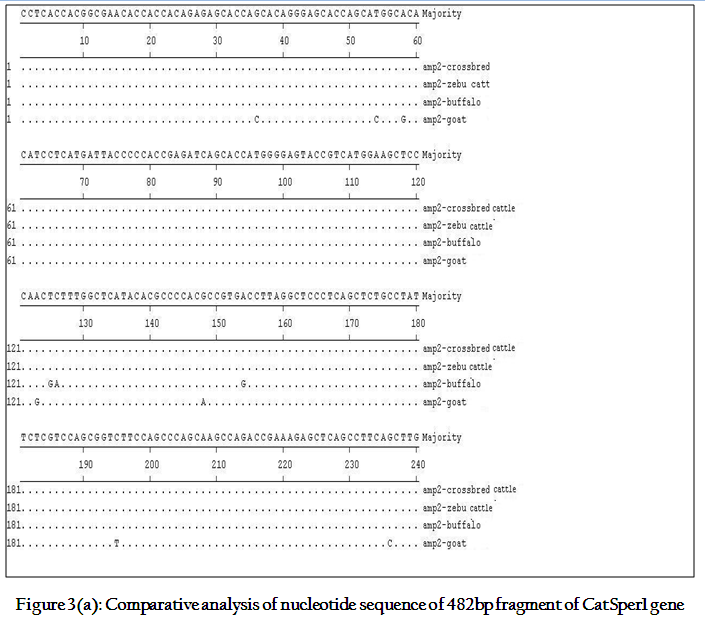
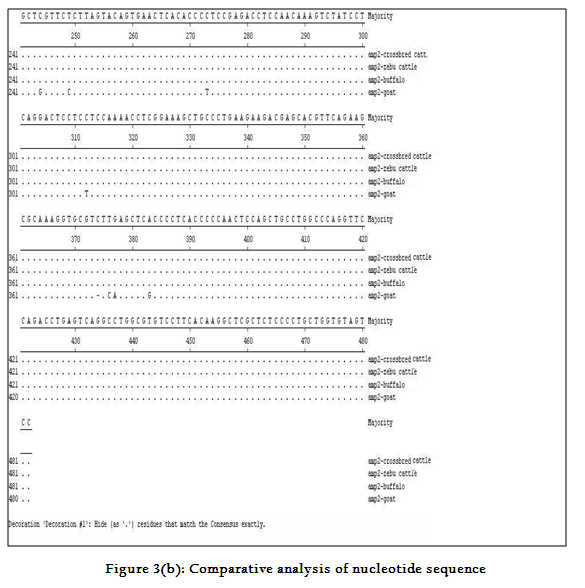
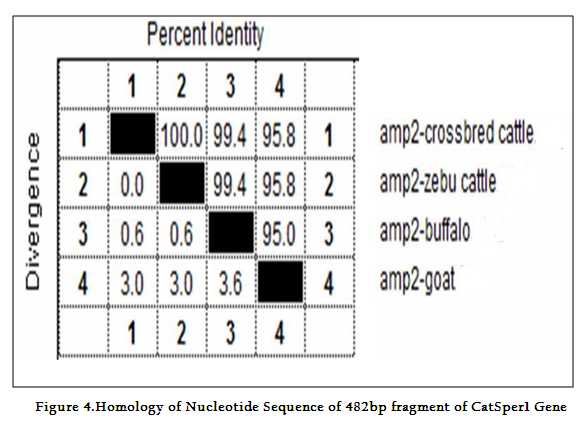
Though the 482 bpamplicon encompassing exon 2 and 3 along with intermediary and flanking introns of CatSper1 gene in crossbred (vrindavani) cattle, zebu (tharparkar), buffalo (murrah), goat (black bengal) weresuccessfully amplified however the sheep (Ovisaries) and mithun (Bosfrontalis) DNA samples could not be amplified. The sequences obtained from these amplicon from these four species (GenBank accession no. JF737759–62) were compared. No variations were noticed between crossbred and zebu cattle. However three nucleotide variations have been noticed in crossbred cattle when compared to buffalo and all were found in intron 3 (Figure 3a, 3b). On comparison of crossbred cattle with goat, three nucleotide variations were observed in the exon 3 and two in intron 3, six in exon 4 and three variations were noticed in intron 4 region. One deletion was witnessed in goat at 374thnucleotide position (Figure 3b)as compared to other ruminants which had a C nucleotide at that position. Three amino acid variations have been noticed in goat. The nucleotide and amino acid sequence homology of crossbred cattle was found to be highest (100%) with zebu cattle (Figure 4).
Poor semen quality is the main cause of high rejection rate in crossbred bulls (Mathew et al., 1982; Rao and Rao, 1991 and Kumar, 2006). Methods have been developed and demonstrated to improve and measure the potentiality of sperm function however importance of candidate gene marker cannot be obviated (Kumar et al. 2013a, 2013b).Genetic characterization and polymorphism identification of CatSper1 is prerequisite for finding a genetic marker of this gene, which may help in improvement of sperm motility and freezability in crossbred cattle. The monomorphic pattern of exon 3 and 4 of CatSper1 gene with respect to different enzymes indicated the conservednessin these exons and suggested to explore polymorphism in other coding regions of this gene as well as other genes responsible for sperm motility. Identification of alleles that are in high association with poor sperm motility in crossbred bulls is the most important area of research now a days and search of a genetic marker in this regard is much needed.
ACKNOWLEDGEMENT
The authors would like to thank the Director, IVRI, Izatnagarand NAIP (Component 4: C30015/4140) for providing financial support and infrastructure.
REFERENCES
Chacon J, Perez E, Muller E, Soderquist L and Martinez HR (1999). Breeding soundness evaluation of extensively managed bulls in Costa Rica. Therio. 52: 221–231.
http://dx.doi.org/10.1016/S0093-691X(99)00124-7
Darszon A, Acevedo JJ, Galindo BE, Hernandez–Gonzalez EO, Nishigaki T, Trevino CL, Wood C and Beltran C (2006). Sperm channel diversity and functional multiplicity. Reprod. 131: 977–998.
http://dx.doi.org/10.1530/rep.1.00612
PMid:16735537
Dhanju CK, Ranjha S, Cheema C and Kaur P (2006).Correlation of Animal spermatozoalfreezability with semen characteristics in crossbred bulls. Ind. J. Anim. Sci.76 (3):241–243.
Geetha T, Kumar S, Dubey PP, Sivamani B, Ghosh SK, Mitra A, Tomar AKS and Sharma A (2011). Sequence variability in CatSper1 gene in Vrindavani crossbred cattle.Ind. J. Anim. Sci.81 (9): 981–983.
Geetha T, Kumar S,Dubey PP, Sivamani B and Sharma A (2013). Polymorphism in Catsper1 Gene in crossbred (Bostaurus X Bosindicus) cattle. Adv. Anim. Vet. Sci. 1 (4): 123–126.
Ghosh SK, Singh SK, Singh LP, Tripathi RP and Tumnyak L (2007).Rejection rate in crossbred bull semen.Compendium XXIII Annual Convention and National Symposium on Challenges in improving reproductive efficiency of farm andpet animals. Bhubaneshwar, Orissa, December 7–9.
Jin JL, O'DohertyAM, Wang S, Zheng H, Sanders KM and Yan W (2005). CatSper3 and CatSper4 encode two cation channel–like proteins exclusively expressed in the testis. Biol. Reprod. 73: 1235–1242.
http://dx.doi.org/10.1095/biolreprod.105.045468
PMid:16107607
Kumar S (2006). Advances in assessment of frozen semen of crossbred bulls. National Seminar on Artificial Insemination: Acceptability, impact, constraints and solutions. New Delhi, India, September 21–14, 2006. pp. 106–116.
Kumar S, Khan S, Sivakumar A, Yathish HM, Sivamani B, Sharma D and Sharma A (2013a). Zona free hamster oocyte penetration test: A reliable in vitro bioassay to assess the bull fertility. Adv. Anim. Vet. Sci. 1 (4): 102–106.
Kumar S, Yathish HM, Sivamani B, Khan S, Sahoo NR, Chauhan A, Sharma D and Sharma A (2013b). Use of platelet activating factor for inducing bull sperm capacitation in vitro: A novel approach.Res.Opin.Anim. Vet. Sci. 3 (10): 348–355.
Lobley A, Pierron V, Reynolds L, Allen L and Michalovich D (2003). Identification of human and mouse Catsper3 and Catsper4 genes: characterization of a common interaction domain and evidence for expression in testis. Reprod. Biol. Endocrinol.1: 53–67.
http://dx.doi.org/10.1186/1477-7827-1-53
PMid:12932298 PMCid:PMC184451
Mathew A, Joseph PJ, Jose TK (1982). Semen characteristics of purebred and cross–bred bulls. Ind. Vet. J. 59: 364–367.
Modi RP, Kumar S, Dubey PP, Sivakumar A, Kumar A, Sivamani B, Ghosh SK, Mitra A and Sharma A (2011). Genetic variability in cation channel of sperm (CatSper1) gene in cattle. National Symposium on Reproductive biotechnologies for augmenting fertility & conservation of animal species with special reference to NEH Region, Aizawl, Mizoram, India, 29 September 27–29, 2011. p. 111
Modi RP, Kumar S, Yathish HM, Dubey PP, Sivakumar A, Sivamani B and Mitra A (2014). Single strand conformation polymorphism in CatSper1 gene in Tharparkarcattle.Ind.J. Anim. Genet. Breed.(In Press).
Qi H, Moran MM, Navarro B, Chong JA, Krapivinsky G, Krapivinsky L, Kirichok Y, Ramsey IS, Quill TA and Clapham DE (2007). All four CatSper ion channel proteins are required for male fertility and sperm cell hyperactivated motility. Proc. Natl.Acad. Sci. U. S. A., 104: 1219–1223.
http://dx.doi.org/10.1073/pnas.0610286104
PMid:17227845 PMCid:PMC1770895
Quill TA, Ren D, Clapham DE and Garbers DL (2001). A voltage–gated ion channel expressed specifically in spermatozoa.Proc. Natl. Acad. Sci. U. S. A. 98: 12527–12531.
http://dx.doi.org/10.1073/pnas.221454998
PMid:11675491 PMCid:PMC60087
Rao BK and Rao AR (1991).Evaluation of crossbred bull for breeding soundness.Ind. J. Anim. Reprod. 12: 111–113.
Ravimurugan T, Kanakaraj P and Thangaraju P (2007).Frozen semen production potential of murrahbulls.Tamilnadu J. Vet. Anim. Sci. 3(5): 269–271.
Ren D, Navarro B, Perez G, Jackson AC, Hsu S, Shi Q, Tilly JL and Clapham DE (2001). A sperm ion channel required for sperm motility and male fertility. Nature., 413: 603–609.
http://dx.doi.org/10.1038/35098027
PMid:11595941
Sambrook J and Russel DW (2001). Molecular cloning– A laboratory manual.3rdedition.Cold Spring Harbor Laboratory Press. New York.
Sharma D, Kumar S, Deb SM, Mitra A, Niranjan SK, Naskar S and Sharma A (2009). Identification of Novel Allelic Variants of Integrin Beta 2 (ITGB2) Gene and Screening for Bubaline Leukocyte Adhesion Deficiency Syndrome in Indian Water Buffaloes (Bubalusbubalis). Anim. Biotechnol. 20 (3): 156 – 160.
http://dx.doi.org/10.1080/10495390902895883
PMid:19544212
Sharma D, Niranjan SK, Kumar S, Deb SM, Naskar S, Sharma A and Mitra A (2010). Molecular characterization of bubaline integrin β2 (ITGB2) cDNA. J. Appl. Anim. Res. 37: 217–220.
http://dx.doi.org/10.1080/09712119.2010.9707127
Sivakumar A, Kumar S, Dubey PP, Yathish HM and Sivamani B (2013a).SNP identification in cation channel of sperm2 (CatSper2) gene in cattle. In National Symposium on "Integrated Development of Vast Biodiversity of Indigenous Livestock for Long Term Rural Livelihood Security" held art o be held at the Department of Animal Genetics and Breeding, College of Veterinary and Animal Sciences, Pantnagar during 7th– 8th February, 2013.
Sivakumar A, Kumar S, Dubey PP, Yathish HM and Sivamani B (2013b). Variability in cation channel of sperm1 (CatSper1) gene in cattle. InNational Seminar on Technological and Policy Interventions of Sustainable Cattle Breeding in India at Project Directorate on Cattle, Meerut on 14th Mar 2013
Tyagi S, Mandal DK, Kumar M and Mathur AK (2006).Reproductive wastage rate of crossbred dairy bulls with reference to level of exotic inheritance and number of breed component.Ind. J. Anim. Reprod. 27(1): 27–30.