Journal of Animal Health and Production
Research Article
Molecular Detection of the Toxin Gene toxA in Pasteurella multocida Isolated from Calves
Zizet Z. Zareh1*, EL Shafei AA2, Shaimaa A Abd Elkader2, Hamada Alazazy2
1Bacteriology Department, Animal Health Research Institute, Dokki, Giza; 2Zagazig Provincial lab. of Animal Health Research Institute.
Abstract | Pasteurella multocida (P. multocida) is frequently found in the upper respiratory tract of domestic animals as an opportunistic agent; it is the main source of an extensive range of sicknesses with universal economic importance and under predisposing management or environmental conditions which establish stress for the livestock such as climate, marketing, transport (shipping fever), change of feed, or ventilation. The present study aimed to isolate and identify the P. multocida organisms in calves and to investigate the antibiotic susceptibility of isolates as well as PCR–based detection of virulence gene (tox A). A total of 168 samples were collected from calves in different farms in Ismailia and El-Dakahlia governorates, Egypt. The samples comprised 124 nasal swabs from apparently healthy and diseased calves and 44 lung samples of calves from diseased emergency-slaughtered and recently dead animals. The bacteriological investigation of the collected samples revealed 11 isolates of P. multocida (6.5%). All isolates of P. multocida were confirmed using the VITEK2 compact system. Antibacterial assay for all the isolates detected the highest sensitivity to ciprofloxacin, gentamicin (100% for each), and tetracycline (81.2%) while it was resistant to amoxicillin, enrofloxacin (100% for each), ampicillin, norfloxacin, (81.8% for each), erythromycin (72.7%), and streptomycin (63.6%). Furthermore, Polymerase Chain Reaction (PCR) assays were used to detect the P. multocida toxA gene. The tested isolates yielded the predicted size for the toxA gene at 864bp. In conclusion, P. multocida (containing toxA) was present in apparently healthy, as well as diseased/ emergency slaughtered calves. The P. multocida isolates were found fully resistant to amoxicillin and enrofloxacin, and susceptible to ciprofloxacin and gentamicin.
Keywords | Pasteurella multocida, calves, VITEK2 Compact system, PCR, toxA gene
Received | December 29, 2020; Accepted | January 19, 2021; Published | March 25, 2021
*Correspondence | Zizet Z Zareh, Bacteriology Department, Animal Health Research Institute, Dokki, Giza; Email: zzakaria2012@yahoo.com
Citation | Zareh ZZ, El Shafei AA, Elkader SAA, Alazazy H (2021). Molecular detection of the toxin gene toxa in pasteurella multocida isolated from calves. J. Anim. Health Prod. 9(s1): 14-19.
DOI | http://dx.doi.org/10.17582/journal.jahp/2021/9.s1.14.19
ISSN | 2308-2801
Copyright © 2021 Zareh et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
INTRODUCTION
Pasteurella multocida (P. multocida) is a Gram-negative bacterium that causes pasteurellosis, an acute septicemic disease with high mortality and substantial losses in livestock (Jakeen et al., 2016) and considered as a critical factor in pneumonia (Haji Hajikolaei et al., 2010). Calves of all ages are affected with P. multocida, however six months to two years age is highly susceptible, representing 14% of respiratory diseases in calves (Radostits et al., 2007). The organism is manifest in animals’ upper respiratory tract as a normal flora; however, stress or compromised immunity makes the bacteria cause severe respiratory issues (Thorbjorg et al., 2016).
Pasteurella multocida causes many sicknesses in livestock all over the world; the most common is Hemorrhagic Septicemia (HS) (Gharibi et al., 2017). Mostly, the disease affects calves within four weeks of weaning when they are sold to various farms. Hence, it is commonly dubbed as the «Shipping fever» (Snowder et al., 2006). P. multocida outbreak showed high fever, respiratory distress, nasal discharge, polypnea, and death across the span of few days. Afterward, severe congestion in the trachea, lung, liver, and small intestine was found (Azizi et al., 2011).
Pasteurellosis diagnosis in livestock still requires clinical manifestations, post mortem findings, and conventional bacteriological approaches that identify the causes. These methods are lengthy and hinder the differentiation between P. multocida and especially the Pasteurella haemolytica (Mannheimia haemolytica) (Berge et al., 2006; Bell, 2008). The VITEK2 technique added the bacterial screening’s field by availing a speedy, economic, and highly sensitive technique for the identification of the bacteria (Wallet et al., 2005). The PCR assay is highly specific and sensitive, providing a swift revelation of P. multocida, without any regard to the samples’ purity. Therefore, only PCR positive samples are the most effective and with a high-quality yield that could avail the isolation of the organism (Kalorey et al., 2008).
Pasteurella multocida strains with various characteristics of virulence show divergent expression of virulence genes (Yan Cheng et al., 2020). Toxigenic and nontoxigenic P. multocida isolates retain the same diagnostic biochemical morphology and reactions. It is necessary to further test laboratory isolates to classify toxigenic and nontoxigenic strains (Nagai et al., 1994).
The toxA gene can be considered as a significant epidemiological marker for the characterization of P. multocida isolates (Aski and Tabatabaei, 2016). The toxA gene is a virulence gene related to P. multocida; is necessary to detect pathogenic P. multocida (Varte et al., 2014). The toxA gene sequence shows that detection assays for this gene, including PCR, are valid for the identification of toxigenic isolates of P. multocida (Buys et al., 1990; Peterson, 1990.
The present study aimed to isolate and identify the P. multocida isolates in apparently healthy, diseased, and recently dead and slaughtered calves using a rapid and accurate technique (VITEK2 compact system) and to monitor the antibiotic susceptibility and to perform PCR–based detection of certain virulence genes (tox A).
MATERIALS AND METHODS
Animals
The study included diseased, dead, and apparently healthy animals at different farms in Ismailia and EL-Dakahlia governorates, Egypt. The majority of the affected cases manifested respiratory infections’ symptoms such as nasal mucopurulent discharge, cough, respiratory manifestations, and fever. The experimental protocols were carried out as per guidelines of Ethics of Animal Use in Research Committee, Faculty of Veterinary Medicine, Zagazig University, Egypt.
Samples
A total of 124 nasal swabs (104 were collected from diseased calves and 20 from apparently healthy ones) and 44 lung samples from disease- emergency slaughtered and recently dead calves were collected. The nasal swabs were collected from both nasal cavities. Then they were displaced in a non-nutritive transport medium, e.g., phosphate-buffered saline, where the temperature was within 4–8ᴼC to stop the spoilage of samples or further over growth of bacteria. Time of transition was within 24 hours. Pneumonic lungs were packed separately in sterile plastic bags, which were then labeled and moved in an icebox to the laboratory. All samples were taken with care and ethical guidelines and biosafety measures were followed during sampling and also during working in the laboratory.
Bacteriological Examination
Nasal swabs were inoculated into a Casein Sucrose Yeast (CSY) broth for 6–8 hours, and then a loopful was cultivated onto CSY, sheep blood, and MacConkeyʼs agar media (Oxoid, UK). Lung specimens were cultured directly onto the previously mentioned media. All agar plates were incubated at 37ᴼC for 48 hours. Suspected colonies were picked up and subjected to morphological and biochemical identification. The biochemical tests included oxidase, indol, catalase, urease production, and nitrate reduction (Oxoid, UK) were performed as described previously (Quinn et al., 2011).
Identification of P. multocida using the VITEK2 Compact System
The P. multocida which identified by conventional method were subjected to further confirmation via the VITEK2 compact system (Biomeriux VITEK-2 Compact ref Manual – Ref-414532- France) according to the instructions of manufacturer (Pincus, 2006) as following:
Suspension preparation: An adequate number of interesting colonies of pure culture were transferred via sterile swabs, where they were suspended in 3.0 mL of sterile saline (pH 4.5–7.0) in a polystyrene (12 x 75 mm) test tube. Then, the mixture was mixed well. The turbidity was adjusted to be the equal of a 0.5–0.63 McFarland turbidity with a VITEK2 instrument DensiChek (BioMerieux, France).
Inoculation: For each isolate, the Gram-negative (GN) documentation cards were inoculated with the microorganism suspension. The isolate was identified using 47 different biochemical tests. A test tube with the suspension of microorganism was placed into a special rack (cassette) which was in turn displaced into a vacuum chamber station. Afterward the application of vacuum and introduction of air into the station, the bacterial suspension forced through the transfer tube into microchannels that fill all the test wells.
Card Sealing and Incubation
Injected cards were passed by means of cutting off the transfer tube and sealing the card before loading into the carousel incubator. All cards were raised on line at 35.5 ͦ C ± 1.0 C for approximately six hours. During incubation, the cards were read every 15 minutes automatically. The results were got automatically and printed within 6–8 hours. All used cards were automatically dispensed into waste containers.
Antimicrobial Susceptibility Testing
All identified bacterial isolates were sub-cultured into the broth that were adjusted to be equivalent to 0.5 MacFarland turbidity, and then swabbed onto Mueller Hinton agar (Oxoid, UK) plates and then left to dry (Quinn et al., 2011). The antibiotic discs were then placed and the plates were aerobically incubated at 37oC for 24 hours. The zones of inhibition around the antibiotic discs were measured and interpreted according to the Clinical Laboratory Standards Institute (CLSI, 2017). The following antibiotic discs were used: amoxicillin (10μg), ampicillin (20μg), ciprofloxacin (5μg), Enrofloxacin (5μg), erythromycin (15μg), gentamicin (10μg), norfloxacin (10μg), streptomycin (25μg), tetracycline (30μg), and trimethoprim-Sulfamethoxazol (25μg) (Oxoid, UK). Drug resistance of antibiotic was calculated with MRA index according to (Rotchell and paul 2016).
Detection of the toxA Gene by PCR
The PCR assay was carried out on the isolated strains of P. multocida (Tang et al., 2009) as following:
DNA Extraction: DNA was extracted from the assessed isolates via the QIAamp DNA Mini kit (Qiagen, Germany, GmbH) as per the manufacturer’s instructions: 200 μL of the isolate suspensions were incubated with 200 μL of lysis buffer and 10 μL of proteinase K for ten minutes at a temperature of 56°C. Then, 200 μL of ethanol with a 100% concentration was introduced to the lysate. The sample was then rinsed and centrifuged as per the manufacturer’s instructions. Nucleic acid was eluted with 100 μL of elution buffer provided in the kit.
PCR Amplification: The toxA amplification occurred in 25μL reaction volume containing Emerald Amp Max PCR Master Mix (12.5 μL; Takara, Japan), 1 μL of each primer (Metabion Germany) of 20 pmol concentration, 6 μl of DNA template and 4.5 μL of water. The reaction occurred in an applied biosystem 2720 thermal cycler. The PCR amplification was done in a thermocycler (Applied Biosystems, USA) using the thermal conditions as initial denaturation at 94°C for ten minutes and 35 cycles of 94°C, 48°C for a minute, 72°C for a minute each, after which a final extension occurred at 72°C for ten minutes (Table 1).
Analysis of the PCR products: The PCR products were separated by electrophoresis on 1% agarose gel (Applichem, Germany, GmbH) in 1x TBE buffer at 40°C using gradients of 5 V/cm. For gel study, 15 μl of the products was loaded in each gel slot. A 100 bp plus DNA ladder (Qiagen, Germany, GmbH) was adopted to know the fragment sizes. The gel was photographed by a Gel documentation system (Alpha Innotech, Biometra) and the information was evaluated via computer software.
RESULTS
Bacteriological investigation of the nasal swabs from apparently healthy calves revealed one positive sample out of 20 nasal swabs with incidence of 5.0%; meanwhile, out of 104 nasal swabs samples of diseased calves revealed six isolates of P. multocida with incidence of 5.8%. Isolates from lung tissue samples from recently dead and emergence slaughtered calves exhibited four positive samples (out of 44) with incidence of 9.1%. A total of 11 (6.5%) P. multocida positive samples were observed in apparently healthy, diseased, and recently dead and emergence slaughtered samples (Table 2). All colonies (11) were reinvestigated using the VITEK2 compact system and the results confirmed isolation of 11 P. multocida isolates with an incidence of 100%. A confirmatory identification by the VITEK2 compact system using Gram-negative cards was employed. The identification cards contained 47 different biochemical tests.
All the P. multocida isolates were sensitive to ciprofloxacin, gentamicin (100% for each), and tetracycline (81.2%) while exhibited multidrug resistance against amoxicillin, and enrofloxacin, 100% for ampicillin, norfloxacin (81.8%), erythromycin (72.7%), streptomycin (63.6%), and trimethoprim- Sulphamethoxazol (54.5%). Multi-antibiotic resistant (MAR) values greater than 0.2 means that the P. multocida had a high resistance to the antibiotics (Table 3).
PCR assay detected toxA gene in all isolates of P. multocida strains showed bands at 864 bp as in (Figure 1).
DISCUSSION
Pasteurella multocida is an important pathogen hazardous to animal and human beings with a worldwide distribution (Hung et al, 2010). It causes various economical diseases in livestock all over the world, among these most common is Hemorrhagic Septicemia (Gharibi et al., 2017). Isolation rates of P. multocida from nasal swabs of apparently healthy calves and lung samples of diseased cases were 5.6% and 9.1%, respectively. The total isolation of P. multocida was 6.5%. Previous studies reported 8.3% (Sedeek and Thabet,
Table 1: Primers sets sequences, target genes, amplicon sizes and cycling conditions.
| Target gene | Primers sequences | Amplified segment (bp) | Primary Denaturation | Amplification (35 cycles) | Final extension | Reference | ||
| Secondary denaturation | Annealing | Extension | ||||||
| toxA | CTTAGATGAGCGACAAGG | 864 |
94˚C 10 min. |
94˚C 1 min. |
48˚C 1 min. |
72˚C 1 min. |
72˚C 10 min. |
Tang et al. 2009 |
| GAATGCCACACCTCTATAG | ||||||||
Table 2: Prevalence of Pasteurella multocida isolated from apparently healthy diseased, dead and slaughtered calves
| Samples | Apparently Healthy calves (n=20) | Diseased calves (n=104) | Dead & Slaughtered calves (n=44) | total (n=168) | |||||
| Type of samples | No. | No | % | No | % | No | % | No |
% |
| Nasal swabs | 124 | 1 | 5.0 | 6 | 5.8 | 0 | 0 | 7 | 5.6 |
|
Lung tissue |
44 | 0 | 0 | 0 | 0 | 4 | 9.1 | 4 | 9.1 |
|
Total |
168 | 1 | 5.0 | 6 | 5.8 | 4 | 9.1 | 11 |
6.5 |
% calculated according to No. of examined sample
Table 3: Antibiogram pattern of Pasteurella multocida isolated from clinically healthy, diseased and recently dead & slaughtered calves
| Antimicrobial agent (potency) | Number of P. multocidaisolates showing Antimicrobial susceptibility patterns (%) | |
| Resistance | Sensitivity | |
|
Amoxicillin (10μg) |
11 (100) | 0 (0) |
|
Ampicillin (20μg) |
9 (81.8) | 2 (18.2) |
|
Ciprofloxacin (5μg) |
0 (0) | 11 (100) |
|
Enrofloxacin (5μg) |
11 (100) | 0 (0) |
|
Erythromycin (15μg) |
8 (72.7) | 3 (27.3) |
|
Gentamicin (10μg) |
0 (0) | 11 (100) |
|
Norfloxacin (10μg) |
9 (81.8) | 2 (18.2) |
|
Streptomycin (25μg) |
7 (63.6) | 4 (36.4) |
|
Tetracycline (30μg) |
2 (18.2) | 9 (81.2) |
|
Trimethoprim-Sulphamethoxazol (25μg) |
6 (54.5) |
5 (45.5) |
2001) to 15.89% (Sayed and Zaytoun, 2009) isolation of P. multocida in Assuit Governorate Egypt, and 15% in England (Defra, 2006). Higher incidence rates of P. multocida were obtained elsewhere in Egypt i.e., 19.23% (Enany et al., 2012) and 22.73% (Khadr, 2005).
The VITEK2 compact system is well known for confirmation of biochemically identified organisms. The confirmatory identification by the VITEK2 compact system was based on GN cards. The identification cards contain 47 different biochemical tests. Concluding results were yielded in roughly 6–8 hours (Pincus, 2006). All tested isolates of P. multocida were confirmed by VITEK2 compact that showed 100% positively of P. multocida. The VITEK2
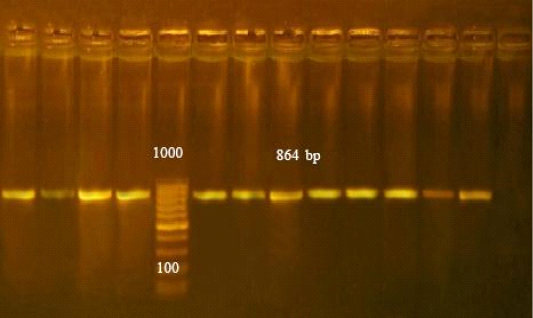
Figure 1: Agarose gel electrophoresis of PCR products of toxA gene of Pasteurella multocida isolates. PCR amplification of toxA gene among the investigated isolates revealed that all isolates were positive for toxA gene with amplified fragments at 864bp. Lane L: 100-1000 bp ladder as molecular size (DNA marker). Lane Pos: Control positive Pasteurella multocida. Lane Neg: Control negative. Lanes from 1 to 11: Positive Pasteurella multocida toxA gene (864 bp).
system poses quite the potential, is a highly automated novel tool for expedited identification of Gram-negative bacilli isolated from clinical specimens (Farid et al., 2014).
Antibiotic utilization in bovine respiratory disease (BRD) treatment is widely used. Multidrug-Resistant (MDR) pathogens related to BRD were observed and deemed as threats to the livestock industry (Klima et al., 2014). Antimicrobial susceptibility should be perpetually monitored to curb MDR strains not that even transforms from livestock to humans (Holman et al., 2015; Klima et al., 2014). In this study, P. multocida was sensitive to ciprofloxacin and gentamicin (100% each) and tetracycline (81.2%). These results agree with (Khamesipour et al., 2014) and (Kumar et al., 2009; Shayegh et al., 2010) who observed 100% sensitivity of the isolated P. multocida to ciprofloxacin. The results agree as well with (Algammal et al., 2020) who reported that all the tested isolates of P. multocida were sensitive to gentamicin, enrofloxacin, and norfloxacin, while it exhibited a multidrug resistance against ampicillin, amoxicillin, erythromycin, and streptomycin (100% each). The results disagree with (Varte et al., 2014) mentioned that the majority of the isolates of P. multocida were 100% sensitive to amoxicillin, cephalexin, chloramphenicol, ciprofloxacin, enrofloxacin, erythromycin, gentamicin, and tetracycline while it exhibited a multidrug resistance against oxytetracycline (93%) followed by ampicillin (87%) and amikacin (47%). In this study, the MAR index is calculated as the ratio of the number of antibiotics to which the organism is resistant to the total number of antibiotics to which the organism is exposed. The MAR index exceeded 0.2 due to the high risk of the used antibiotics. The antimicrobial susceptibility of P. multocida is important to determine resistance development and help pick the appropriate antibiotics against the diseases caused by P. multocida.
Virulence factors help the pathogen colonize and invade the host, avoid defense mechanisms, injure tissues, and stimulate inflammatory responses (Ewers et al., 2004).
The toxA gene is a virulence gene associated with P. multocida was identified by PCR in all tested isolates. Earlier research observed that PCR targeting the toxA gene is sensitive and crucial for pathogenic P. multocida detection (Varte et al., 2014). In the present study, all P. multocida strains showed bands at 864 bp (Figure 1). These results agree with Lichtensteiger et al. (1996). Many authors, (Ewers et al., 2006; Shayegh et al., 2010; Khamesipour et al., 2014) recorded toxA genes in P. multocida isolates. The toxA gene is not inserted into the bacterial chromosome but in a lysogenic bacteriophage that infects the agent (Pullinger et al., 2004) toxA plays an important role in destruction of lung tissues, and stimulation of a noxious host inflammatory response. Therefore, the toxA gene is considered an epidemiological marker found mostly in pneumonic P. multocida isolates (Harper et al., 2006).
CONCLUSION
From the results it could be concluded that P. multocida (containing toxA) was present in clinically healthy, diseased and recently dead or slaughtered calves in Ismailia and EL-Dakahlia governorates, Egypt. All isolates were found 100% resistant to amoxicillin and enrofloxacin, and susceptible to ciprofloxacin and gentamicin.
conflict of interest
There is no conflict of interest.
authors contribution
All authors contributed equally.
REFERENCES





