Journal of Animal Health and Production
Research Article
Occurrence of Escherichia coli as a Causative Agent of Enteritis in Dogs with Special Reference to Their Multidrug Resistance and Virulence Genes
Zizet Z. Zarea1*, Ghada O. EL-Demerdash2, EL Shafei A. A.3, Shaimaa A. Abd Elkader3
1Bacteriology Department, Animal Health Research Institute, Dokki, Giza; 2AL Fayoum Provincial laboratory of Animal Health Research Institute; 3Zagazig Provincial laboratory of Animal Health Research Institute.
Abstract | Escherichia coli (E. coli) massively causes deaths all around the world. Human–pet proximity may inadvertently harm humans. Dogs are susceptible to virulent E. coli strains causing enteric infections in humans. In the present study, a total of 90 rectal swabs were collected from dogs of different ages (suffered from diarrhea with fever, nausea, chills, loss of appetite, and bloating) at different veterinary hospitals and clinics in Cairo to determine the incidence rate of E. coli and their virulence and resistance genes. Bacteriological examination revealed an overall E. coli occurrence rate of 67.7% (61/90). The highest isolation rate of 75.5% was from puppies that were 3 months and more. Serotyping of ten E. coli isolates showed that they belonged to seven serogroups O18, O27, O55, O126, O148, O158, and O166 and other untypable three strains. All E. coli isolates were subjected to disc diffusion sensitivity tests and were resistant to tetracycline, trimethoprim/ sulphamethoxazole (100% for each), cefotaxime (95.1%), and erythromycin (93.4%), while they were sensitive to amikacin only (88.5%). The occurrence of virulence genes in the seven tested E. coli isolates was conducted using PCR. It was revealed that the stx1 gene was detected in O18, O126, O148, O158, and O166 serogroups while the stx2 gene was detected in O18 and O27 only. The eaeA gene was detected in O27, O55, O148, and O158 serogroups. Also, the antibiotic-resistant blaTEM and tetA genes were detected in all the seven tested serotypes. These combined results indicate that pet animals may harbor E. coli causing diarrhea at different ages.
Keywords | Diarrhea, Dogs, Enteropathogens, Escherichia coli, Virulence genes, Shiga toxin, Drugs resistance.
Received | December 29, 2020; Accepted | January 19, 2021; Published | March 25, 2021
*Correspondence | Zizet Z Zareh, Bacteriology Department, Animal Health Research Institute, Dokki, Giza; Email: zzakaria2012@yahoo.com
Citation | Zarea ZZ, El-Demerdash GO, El-Shafei AA, Abd Elkader SA (2021). Occurrence of escherichia coli as a causative agent of enteritis in dogs with special reference to their multidrug resistance and virulence genes. J. Anim. Health Prod. 9(s1): 7-13.
DOI | http://dx.doi.org/10.17582/journal.jahp/2021/9.s1.7.13
ISSN | 2308-2801
Copyright © 2021 Zareh et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Escherichia coli (E. coli) usually inhabits the gastrointestinal tract of animals; it can affect dogs, and humans. Also, it can be transmitted from animals to humans and vice versa. They are one of the most common enteric types present worldwide. Under certain conditions, these bacteria can also be responsible for causing diseases. Dogs and humans can be infected with E. coli by consuming contaminated water or food. E. coli usually affects weak animals, such as the too young, old, and malnourished animals or immune-compromised pets (Hustom, 2019). Canine puppies younger than one year old are quite prone to gastrointestinal infections, with acute diarrhea manifesting as the most usual clinical indicators; it could lead to severe dehydration and death (Hubbard et al., 2007).
Enterotoxigenic E. coli, for instance, is an Escherichia coli type that may induce foodborne sickness, beginning to manifest when compromised water or food is consumed. Enterotoxigenic E. coli emits a toxin that affects the inside of the infected intestinal tract, consequently causing diarrhea (Hustom, 2019). Virulence factors of pathogenic E. coli includes Shiga toxin-production (Croxen et al., 2013). Shiga toxins, stx1 and stx2 produced by the Shiga toxin-producing E. coli (STEC) strains are the chief virulence aspects. The strains possess the LEE (locus for the enterocyte effacement), where the eae gene resides. This gene encodes for intimin, the adherence protein whose receptor is required for virulence (Ben Said et al., 2019). The ability of STEC to induce serious diseases correlates to the emittance of one or more Shiga-like toxins) stx1, stx2, or other variants); they inhibit the host cells’ synthesis of protein, leading to cell death (Ahsan et al., 2020).
Dogs that carry multidrug-resistant E. coli strain (MDREC) in their feces could contaminate the surrounding environment, as a result of transmitting these bacteria to humans as well as other animals (Warren et al., 2001). E. coli is often prone to various antibiotics; however, by time and antibiotics’ excessive use, drug-resistant strains emerged. Moreover, extended-spectrum β-lactamases (ESBL) that induce enteric pathogens is a serious problem (Mathur et al., 2002).
This work aims to detect the occurrence rate of E.coli in diarrheic dogs of different ages, identify the isolates by VITEK2 compact, serological testing, and serogroup, as well as investigate the presence of the virulence (stx1, stx2, and eae) and drug resistance (bla TEM and tet A) genes.
MATERIALS AND METHODS
Samples Collection
A total of 90 rectal swabs were collected from dogs of different ages (one month to over one year) from different veterinary hospitals and clinics in Cairo. The animals were suffered from diarrhea with fever, bloating, chills, loss of appetite, and nausea. Each sample was collected using a sterile swab that was kept in an icebox immediately after collection and then sent to the laboratory without delay. All samples were taken with care about ethical guideline for sampling and also during working all biosafety measures were done. The experimental protocols were conducted according to guidelines of Ethics of Animal Use in Research Committee, Faculty of Veterinary Medicine, Zagazig University, Egypt.
Isolation and Identification of E. coli
Rectal swabs were cultivated into nutrient broth (Difco) tubes and incubated aerobically at 37°C for 18 hours. A loopful from each broth culture was inoculated onto MacConkey agar (Oxoid) (Cruickshanak et al., 1975) and Eosin Methylene Blue (EMB) agar (Oxoid) (Koneman et al., 1988). Agar plates were incubated aerobically for 24 hours at 37°C and then were observed to detect if any bacterial colonies were present. Suspected colonies were assessed for Gram`s reaction. Gram-negative bacilli colonies were found. Each sample was further subjected to biochemical testing such as catalase, urease test, Triple Sugar Iron agar (TSI), methyl red test, indole test, citrate test, and Voges-Proskauer (IMViC) (McFaddin, 1985) as the traditional method of identification. Then, results were confirmed by VITEK2 compact identification and serotyping.
VITEK2 Compact System Identification
Identification by VITEK2 compact system was done according to the manufacture’s instruction (Biomeriux VITEK-2 Compact ref Manual – Ref-414532- France) (Pincus, 2006).
Serological Identification
Ten E. coli isolates were submitted to slide agglutination test via polyvalent antisera per instruction of manufactures procedures (E. coli antisera Denka Seiken Co. LTD) (Collee et al., 1996).
Antimicrobial Susceptibility Testing
Fresh cultures of all isolates of E. coli were tested for antimicrobial susceptibility via the standard disk diffusion method as per guidelines of the Clinical Laboratory Standards Institute (CLSI, 2017). The following antimicrobial discs were used: amikacin (AK, 30 μg), amoxicillin/clavulanic acid (AMC, 20/10 μg), ampicillin (Amp, 15 μg), cephalothin (KF, 30 μg), ciprofloxacin (CIP, 30 μg), doxycycline (DO, 30 μg), erythromycin (E, 5μg), neomycin (N, 30 μg), tetracycline (TE, 30 μg), and trimethoprim/sulphamethoxazole (SXT, 25 μg) (Oxoid).
Detection of Some Virulence and Antibiotic Resistance Genes of E. coli by PCR
DNA was extracted from the seven E. coli serotypes to detect virulence and antibiotic resistance genes via the QIAamp DNA Mini kit (Qiagen, Germany, GmbH) according to the manufacturer’s recommendations. 200 µl of the sample suspension was incubated with 200 µl of lysis buffer and 10 µl of proteinase K at 56OC for 10 minutes. Afterward, 200 µl of ethanol (100%) was added to the lysate. The sample was rinsed and centrifuged as per the instructions of the manufacturer. Nucleic acid was eluted with 100 µl of elution buffer found in the kit. The used primers sets were procured from Metabion (Germany). Table 1 illustrates the cycling parameters.
Primers were applied in a 25- µl reaction mixture of 12.5 µl of Emerald Amp Max PCR Master Mix (Takara, Japan), 1 µl of each 20 pmol primer concentrations, 6 µl of DNA template and 4.5 µl of water.
PCR outcomes were dispersed by electrophoresis on 1.5% agarose gel (Applichem, Germany, GmbH) in 1x TBE buffer at room temperature utilizing gradients of 5V/cm. For gel examination, 20 µl of the PCR products were loaded in each gel slot. Gelpilot 100 bp DNA Ladder (Qiagen, Germany, GmbH) was adopted to know the fragment sizes. A gel documentation system (Alpha Innotech, Biomet
Table 1: Primers sequences, target genes, amplicon sizes, and cycling conditions of E. coli
| Target gene | Primers Sequences | ze (bp) | Primary Denaturation | Amplification (35 cycles) | Final Extension | Reference | ||
| Secondary Denaturation | Annealing | Extension | ||||||
| eaeA | F: ATG CTT AGT GCT GGT TTA GG |
248 |
94˚C 5 min. |
94˚C 30 sec. |
55˚C 30 sec. |
72˚C 30 sec. |
72˚C 7 min |
Bisi-Johnson et al., 2011 |
| R: GCC TTC ATC ATT TCG CTT TC | ||||||||
| stx1 | F:ACACTGGATGATCTCAGTGG |
614 |
94˚C 5 min. |
94˚C 30 sec |
58˚C 40 sec |
72˚C 45 sec |
72˚C 10 min |
Dipineto et al., 2006
|
| R:CTGAATCCCCCTCCATTATG | ||||||||
| stx2 | F:CCATGACAACGGACAGCAGTT |
779 |
||||||
| R:CCTGTCAACTGAGCAGCACTTTG | ||||||||
| tetA | F:GGTTCACTCGAACGACGTCA |
576 |
94˚C 5 min |
94˚C 30 sec |
50˚C 40 sec |
72˚C 45 sec |
72˚C 10 min |
Randall et al. 2004 |
| R:CTGTCCGACAAGTTGCATGA | ||||||||
| blaTEM | F;ATCAGCAATAAACCAGC | 516 |
94˚C 5 min |
94˚C 30 sec |
54˚C 40 sec |
72˚C 45 sec |
72˚C 10 min |
Colom et al., 2003 |
ra) was used to get the gel photograph. Computer software was utilized to analyze the data.
RESULTS
Out of the 90 examined rectal swabs, 61 were positive for E. coli (67.7%) at different ages. The highest isolation rate was in puppies of less than three months old and recorded as 75.5% and the lowest frequency was 57.1% was recorded from dogs of more than one year old as shown in Table 2.
Ten isolates were representing seven serogroups including O18, O27, O55, O126, O148, O158, and O166 in addition to 3 untypable groups.
Table 2: Occurrence of E. coli in diarrheic dogs of different ages,
| Age of Dogs | No. of Examined Samples | No. of E. coli Positive samples | ||
| No. | % | |||
| ≤ 3 months | 53 | 40 | 75.5 | |
|
˃ 3 months – 6 months |
18 | 11 | 61.1 | |
|
˃ 6 months–≤ 9 months |
12 | 6 | 50.0 | |
| Over one year | 7 | 4 | 57.1 | |
| Total | 90 | 61 | 67.7 | |
All isolates of E. coli subjected to disc diffusion sensitivity test showed high drug resistances levels (100%) was against tetracycline and trimethoprim/ sulphamethoxazole, followed by cephalothin, (95.1%) and erythromycin (93.4%), while the highest sensitivity (88.5%) was to amikacin (Table 3).
Table 3: Antimicrobial susceptibility of E. coli isolates (n=61)
| Antibiotics | Resistant Isolates | Sensitive Isolates | ||
| % | No. | % | No. | |
| Tetracycline (TE) | 100 | 61 | 0 | 0 |
| Trimethoprim/Sulphamethoxazole (SXT) | 100 | 61 |
0
|
0 |
| Cephalothin (KF) | 95.1 | 58 | 4.9 | 3 |
| Erythromycin (E) | 93.4 | 57 | 6.6 | 4 |
| Ciprofloxacin (CIP) | 90.2 | 55 | 9.8 | 6 |
| Doxycycline (DO) | 83.6 | 51 | 16.4 | 10 |
| Amoxicillin/Clavulanic Acid (AMC) | 82 | 50 | 18 | 11 |
| Ampicillin (Amp) | 59 | 36 | 41 | 25 |
| Neomycin (N) | 32.8 | 20 | 67.2 | 41 |
| Amikacin (AK) | 11.5 | 7 | 88.5 | 54 |
The E. coli virulence genes of the tested isolates showed amplicons of 779, 614, and 248 bp for stx1, stx2, and eae genes, respectively. Of the seven-tested E. coli isolates (seven serogroups) five strains were positive to stx1 gene, two strains to stx2 gene, and four strains to eae gene (Table 4 and Figures 1 and 2). While the bla TEM and tetA antibio-
Table 4: Detection of virulence and multidrug resistance genes in E. coli serotypes isolated from diarrheic dogs.
| Strain No. | Serogroup |
|
Virulence Genes | Drug-Resistant Genes | |||
|
|
|
eaeA | stx2 |
stx1 |
tetA | blaTEM | |
| 1 | O27 |
|
+ | + |
- |
+ | + |
| 2 | O166 |
|
- | - |
+ |
+ | + |
| 3 | O126 |
|
- | - |
+ |
+ | + |
| 4 | O158 |
|
+ | - |
+ |
+ | + |
| 5 | O148 |
|
+ | - |
+ |
+ | + |
| 6 | O18 |
|
- | + |
+ |
+ | + |
| 7 | O55 |
|
+ | - |
- |
+ | + |
(Absent); + (Present)
tic-resistant genes representing 516 and 576 bp size, respectively, were detected in all the seven tested isolates (seven serogroups) (Table 4 and Figures 3 and 4).
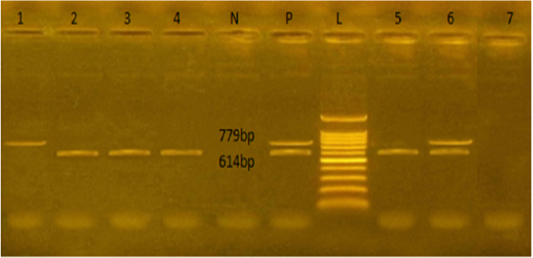
Figure 1: Agarose gel electrophoresis showed PCR products of E.coli stx1and stx2 genes amplified at 614 and 779 bp respectively from seven isolates. L: representing the molecular size marker (100 pb plus ladder). Lane 1 was positive to stx2, lanes 2,3,4, and 5 were positive to stx1. Lane 6 was positive to stx1 and stx2, and lane 7 was negative to stx1 and stx2. N: represents control negative, P: control positive
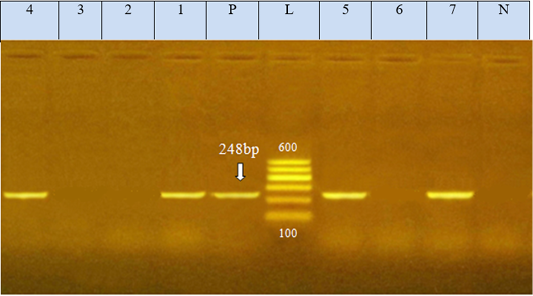
Figure 2: Agarose gel electrophoresis showed PCR product of E.coli eae gene amplified at 248 bp from seven isolates, N: represents negative control; P: represents positive control of eae gene (248 bp). L: represents the molecular size marker (100pb plus ladder). Lanes 1, 4, 5, and 7 were positive to eae gene. Lanes 2, 3, and 6 were negative to eae gene
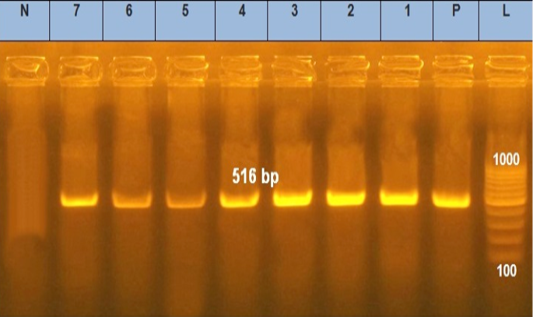
Figure 3: Agarose gel electrophoresis showed PCR products of E .coli blaTEM. Lane L was within 100–1000 bp DNA ladder. N: negative control; P: positive control of blaTEM gene 516 bp. Lanes 1–7 were positive to bla TEM gene
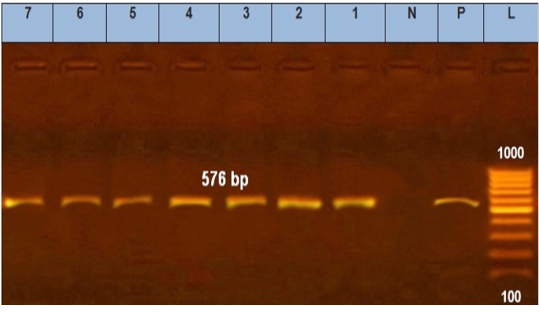
Figure 4: Agarose gel electrophoresis showed PCR products of E. coli tetA. N: represents negative control. P: represents positive control of tetA gene (576 bp). Lane L was within 100–1000 bp DNA ladder (1–7) positive to tetA gene.
DISCUSSION
Dogs and puppies are one of the critical carriers of E. coli, one of the chief causatives of diarrhea and other diseases in humans (Hasan et al., 2016). Out of the 90 examined rectal swabs, 61 were positive for E. coli (67.7%) at different ages. The highest isolation rate was in puppies of less than three months old and recorded as 75.5% and the lowest frequency was 57.1% was recorded from dogs of more than one year old.
In this study, E. coli was isolated from diarrheic dogs at different ages, with total isolation percentages of 67.7% (61/90). Puppies under three months old were the most susceptible to E. coli infection; 75.5% of this pool agrees with Broes et al. (1988) who reported that E. coli was present in diarrheic puppies from one month to one month and a half old. Dogs older than one year were less susceptible to E. coli infection (Table 2). This conforms to various studies exhibiting that the incidence peak of enteritis always occurred within few days after the inception of the weaning period (one month) and the majority of EPEC infections take place in the first three months of lifespan as mentioned by Janke et al. (1989).
From all suspected isolates proved by traditional bacteriological examination to be E. coli, ten random isolates were reinvestigated using the VITEK2 compact system via Gram-negative cards and the results confirm the isolation of E. coli with an incidence of 100%. The VITEK2 compact system is a highly automated new tool for accurate and rapid identification of E. coli. The ten random isolates were subjected to serotyping and the results showed that these ten isolates were representing seven serogroups including O18, O27, O55, O126, O148, O158, and O166 in addition to 3 untypable groups.
On contrary, in other studies in Trinidad, Adesiyun et al. (1997) reported that the prevalence of EPEC strains was not significantly associated with age. Moreover, Marjanca et al. (2002) isolated 24 strains of hemolytic E. coli from dogs with diarrhea. The detected serogroup detected in this study were O18, O27, O55, O126, O148, O158, and O166. On the other hand, Maniam et al. (2020) reported that the most prevalent E. coli serogroup isolated from dogs from feces of healthy ones were O104:H4 and O102:H18. Also, Michele et al. (2014) found that E. coli O145: H 28 was isolated from stray dogs.
All isolates of E. coli subjected to disc diffusion sensitivity test showed high drug resistances levels (100%) was against tetracycline and trimethoprim/ sulphamethoxazole, followed by cephalothin, (95.1%) and erythromycin (93.4%), while the highest sensitivity (88.5%) was to amikacin only (Table 3).
Antimicrobial resistance is constantly evolving since the inception of antibiotics. Many aspects increase bacterial resistance such as disregarding treatment regimen, prophylactic consumption of antibiotics, and using antibiotics as growth promoters (Grave and Tanem, 2006). In our study, many strains showed multiple resistances to tetracycline, and trimethoprim/sulphamethoxazole (100%), cephalothin (95.1%), and erythromycin (93.4%), while the highest sensitivity (88.5%) was to amikacin. This agrees with the results obtained by Younis et al. (2015) that showed high resistance to trimethoprim/sulfamethoxazole (98%), erythromycin (97%), and cefotaxime (95%), while there was high sensitivity to amikacin (95%). Guardabassi et al. (2004) exhibited high sensitivity to amikacin. Habib et al. (2016) reported that E. coli was moderately resistant to tetracycline (54.33%), gentamycin (49.60%), Vibramycin (46.45%), ceftriaxone (44.88%), kanamycin (52.75%), ampicillin (59.65%), ciprofloxacin (25.98%) and norfloxacin (30.70%). These reports and the current study’s findings indicate exponentially increasing resistance patterns of E. coli. It may also imply that E. coli in pet animals transmitted from human represent a G-directional public health hazard (Patoli et al., 2010).
The E. coli virulence genes of the tested isolates showed amplicons of 779, 614, and 248 bp for stx1, stx2, and eae genes, respectively. Of the seven-tested E. coli isolated, seven serogroups: five strains were positive to stx1 gene, two strains to stx2 gene, and four strains to eae gene. While the bla TEM and tet A antibiotic-resistant genes representing 516 and 576 bp size, respectively, were detected in all the seven tested isolates (seven serogroups).
In this study, virulence genes have been detected in E.coli isolated of dog origin where the stx1 and stx2 genes were detected in five E. coli serogroup (O166, O126, O158, O148, and O18) and two E. coli groups (O27 and O18), respectively. stx 1 and stx 2 genes maintain the same pattern in the other reports (Staats et al., 2003; Nakazato et al., 2004; Bentancor et al., 2007).
Herein, the eaeA gene was detected in four strains of different serogroups (O27, O185, O148, and O55). Similarly, Nakazato et al. (2004) reported the eae-positive isolates related to diverse serogroups, including O111:H25, O119:H2, and O142:H6. One E. coli (O18) strain in our work harbored both stx1 and stx2 genes. These results nearly agreed with the findings of Barkalita et al. (2016) who detected both stx1 and stx2 genes in O43 and O130 strains.
In our results, the blaTEM gene was detected in seven tested isolates by PCR at 516 bp, which agrees with the earlier findings (Younis et al., 2015; Delmani et al., 2017). The blaTEM gene found in a plasmid showed a strong correlation between the genogroup conferred resistance determined by PCR and antibiotic susceptibility patterns.
Moreover, tetA gene was found in seven tested isolates at 576 bp amplicon which agrees with (Nde and Logue, 2008) and (Torkan et al., 2015) who suggested that tetA could be the predominant tetracycline gene of resistance harbored by pathogenic E. coli present in dogs in Iran. There are other tetracycline-resistant genes thought to induce resistance via enzymatic inactivation and ribosomal protection.
CONCLUSION
Virulence genes of E. coli isolated from dogs were stx1, stx2, and eae, detected in current study, which probably influence the E. coli enteritis pathogenicity. Moreover, multidrug resistance genes were blaTEM and Tet A detected in E. coli isolates. The results suggested the pet owners to adopt hygiene practice to prevent infection, such as proper food-preparing methods, avoiding drinking water from potentially contaminated sources, thoroughly cooking meat before feeding dogs, and washing hands frequently and thoroughly.
acknowledgements
The authors would like to extend their thanks to all the stuff members at bacteriology unite, animal research institute (AHRI), Egypt for their kind support during this work.
conflict of interest
The authors declare that they no conflict of interest.
authors contribution
Z.Z and A.A planed the work. All authors conducted the practical part of the work and analysed the data. All authors contributed to the article and approved the submitted version.
REFERENCES





