Journal of Animal Health and Production
Review Article
A Complex Genetic Diversity of Newcastle Disease Virus (Ndv) In Africa Continent: An Updated Review
Mohamed Megahed1, Wala Mohamed2, Ola Hassanin1*
1Department of Avian and Rabbit Medicine, Zagazig University, Zagazig, 44511, Egypt; 2The Educational Veterinary Hospital Zagazig University.
Abstract | Newcastle disease (ND) is a highly contagious viral disease of domestic and wild birds with worldwide distributions that listed A by OIE as it causes severe economic losses in the poultry industry. In developing country, it considers a major limiting factor for poultry production which represent an important source for income and food security. In Africa, Newcastle disease virus (NDV) outbreaks is rampant for decades, however the information about the genetic characteristics of the virulent strains circulating Africa is still scarce. Based on the full genome length and F gene sequence, NDV strains are classified into class I (9 genotypes) and II (18 genotypes) within a single serotype. Outbreak in North African countries (like Egypt) caused by genotypes II, VI and VII. In the Eastern African countries such as Tanzania, genotypes V, VII and XIII are the circulating strains. In the central (Nigeria, Niger, Cameroon and Uganda) and western African countries (Mali, Mauritania, Côte d’Ivoire and Burkina Faso), newly circulating genotypes (XIV,XVII and XVIII) are isolated and restricted to this area in addition to other genotypes such as II, VII and V. In Southern African countries, (namely South Africa, Madagascar and Mozambique) genotypes II, VII, VIII, XI and XIII are prevalent. The variable NDV genotypes are been introduced to the different African countries via variable ways, wild and exotic birds, illegal poultry trading through neighboring borders or live bird market. The complex genetic diversity among circulating genotypes, sub optimal prevention afforded by the genotype II vaccine may be major factors that complicate the control of NDV in Africa.
Keywords | Newcastle disease virus, Genotypes, Africa, Genetic diversity, ND
Received | November 17, 2020; Accepted | December 01, 2020; Published | December 27, 2020
*Correspondence | Ola Hassanin, Department of Avian and Rabbit Medicine, Zagazig University, Zagazig, 44511, Egypt; Email: olahassanin@zu.edu.eg, olafalcon2001@yahoo.com
Citation | Megahed M, Mohamed W, Hassanin O (2020). A complex genetic diversity of newcastle disease virus (ndv) in africa continent: an updated review. J. Anim. Health Prod. 9(s1): 97-109.
DOI | http://dx.doi.org/10.17582/journal.jahp/2020/9.s1.97.109
ISSN | 2308-2801
Copyright © 2020 Hassanin et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
Poultry industry is a crucial component of animal production industry, with a huge number of backyard flocks, particularly in the developing countries. They depend on poultry production in the backyard system, to meet household food requirements and as extra-added amount of money source (Maqbool, 2002). Unfortunately, the backyard production system involves a low standard of biosecurity measures and high risk of contagious disease transmission, such as Newcastle disease (ND) (Canan et al., 2012). ND is a highly contagious disease with great negative impact to the poultry industry. Therefore, it is classified as one of the ‘‘notifiable diseases’’ on the World Organization for Animal Health (OIE). Recurrent outbreaks of ND occur across the globe, particularly in the low-income countries.
Newcastle disease virus (NDV), avian paramyxovirus serotype 1 (APMV-1), belongs to the family Paramyxoviridae; genus Avulavirus (Lamb and park, 2007; Samal. 2011; Kuhn et al., 2019). It is an enveloped virus with a non-segmented, single-stranded, negative sense RNA. The virus genome encodes eight proteins, fusion (F) protein, hemagglutinin-neuraminidase protein (HN), nucleocapsid protein (N), phosphoprotein (P), matrix protein (M), large polymerase protein (L), and two additional non-structural proteins, V and W, that is expressed by RNA editing of P mRNA (Lamb and park, 2007). Relying on their pathogenicity in chickens, there are three main pathotypes for NDV isolates, lentogenic (low virulence), mesogenic (intermediate virulence) and velogenic (high virulence) (Beard and Hanson, 1984; Alexander, 1991). The disease in chickens is associated with respiratory distress with often nervous system disorder as well as gastrointestinal and reproductive troubles (Alexander, 1997; Nanthakumar et al., 2000; Tiwari et al., 2004).
Despite the intense vaccination program is being exercised by the poultry breeders across the world, ND has challenged all the rationality and continues to evolve to create new genotypes and sub genotypes that continue to spread across the globe. The current commercially available ND vaccines do not completely induce sterilized immunity as there is still virus shedding and disease can spread in to non-immunized bird (Xiao et al., 2012). Furthermore, most of the commercially available ND vaccines belong to genotype II and are not entirely compatible and deliver complete protection against the newly emerged ND virus species, particularly VII (Kapczynski and King, 2005; Miller et al., 2007; Perozo et al., 2008; Kilany et al., 2015).
NDV has a high impact in large and smallholder African poultry breeders, where it results in negative socioeconomic impact (Alders and Pyme, 2009). They regularly regarded ND as the most significant disease of chickens in Africa. As a disease reportable to OIE, it also affects international trade and national movement of poultry (Alders, 2001). In a 12 year study (January 2000–December 2011) conducted to examine the frequency of recurrent ND outbreaks in 54 African countries, only 40.7 % was discovered to have always reported to OIE (Gardner and Alders, 2014).
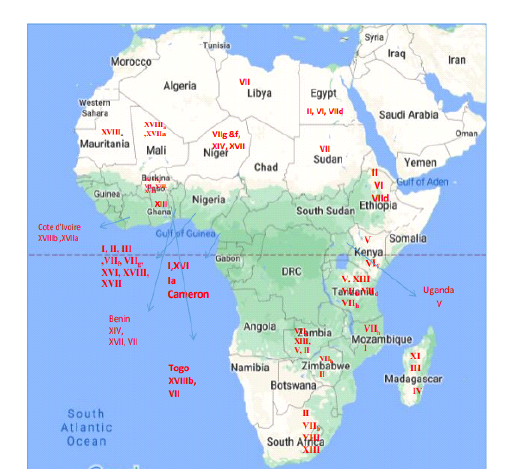
In this review, we decided to state the history and the complex situation of NDV genotyping in Africa. Hence, we focused of the endemic genotypes in each region and their transmissibility across the continent. We presented different reference NDV strains of different genotypes from the Africans continent in Table 1 and Figure 1.
History
The first outbreaks of ND occurred in Indonesia (Java) during 1926 (Kraneveld, 1926) and the same outbreak occurred at a different geographical location on the map of the world, Newcastle-upon Tyne, the UK (Doyle, 1927). However, closely related disease outbreaks in Central Europe have been reported earlier than these dates (Halasz, 1912). One example, Macpherson (Macpherson, 1956) explains the reason of chicken death in the Western Isles of Scotland during 1896 because of NDV infection. Therefore, ND may exist earlier than 1926, but its identification as a specific disease with viral etiology dates back to the outbreaks during this year in Newcastle-upon-Tyne (Alexander et al., 2004).
In the African continents, they were initially diagnosed NDV for the first time during 1944 in South Africa, after chicken’s affections with severe respiratory, nervous and intestinal troubles with high mortalities in the province of Natal (Kaschula et al., 1945). They used in diagnosis serum neutralization test where it was performed at Weybridge laboratory, England. In fact, Kashscula and colleagues believed that the symptoms and post-mortem findings in Natal were so closely to those described by Hudson in Mombasa, Kenya in 1935. Therefore, they suggested that the disease may introduced via ship arrived to Mombasa harbor on the East Coast of Africa. Hudson considered that the infection had spread south to Lindi and the entire African East Coast may be affected (Kaschula et al., 1945). In Madagascar, ND was detected in 1946 (Rajaonarison, 1991), and is now responsible for the mortality rate in the backyard farms, which represent a large sector of Madagascar avian production. In Egypt, ND was identified for the first time in 1948 (Daubney and Mansy, 1948) and since then, Egypt has been regarded as an endemic country by ND. Two years later, they detected the virus in the middle of Congo and the Gambia on the West Coast of Africa (Hamilton, 1950; Lindley, 1951). One year later, it appeared on the Gold Coast (Sudan) and the southern Cameroons (Anon, 1951). In Zambia, they firstly reported the disease in 1952 as the infection spreads along the line of rail with the largest concentration of birds occurred. There was a definite correlation between the number of outbreaks and the amount of vaccine used in any one year (Sharma et al., 1986). Interestingly, due to the presence of the disease
Table 1: NDV reference strains genotypes on Genbank that isolated from different parts in Africa.
| References | Genotype | Accession number |
Strain |
| Snoeck et al; 2013 | I | HF969159 | chicken/Cameroon/CAE11-855/2011 |
|
Snoeck et al; 2009 |
I | HF969159 | chicken/Cameroon/CAE11-855/2011 |
|
Snoeck et al; 2009 |
I | FM200800 | chicken/Nigeria/SH11/2005 |
|
Snoeck et al; 2009 |
I | M24693 | fowl/Australia/Queensland-V4/1966 |
|
Snoeck et al ;2009 |
I | HQ702462 | chicken/Mozambique/ndv42_564/2005 |
|
Snoeck et al; 2009 |
I | AY175774 | pigeon/South Africa/PZAPI99091/1999 |
|
Aldous et al., 2003 |
I | AY 175774 | AV 888/99 MZ-5-99South Africa |
|
Mohamed et al., 2011 |
II | FJ969395 | NDV/chicken/Egypt/4/2006 |
|
Mohamed et al., 2011 |
II | FJ969393 | NDV/chicken/Egypt/2/2006 |
|
Radwan et al ,2013 |
II | JX193769 | NDV/chicken/Egypt/MR2-1998 |
|
Aldous et al .2003 |
II | AY 175708 | AV 72/95 B/K) Zambia |
|
Aldous et al .2003 |
II | AY 175661 | AV 1141/98 10603 South Africa |
|
Aldous et al .2003 |
II | AY 135740 | AV 1433/95 D1702-95 South Africa |
|
Aldous et al .2003 |
II | AY 175710 |
AV 862/95 5249Zimbabwe |
|
Aldous et al .2003 |
II | AY 135755 | AV 992/94 0915/94South Africa |
|
Fenti et al 2013 |
II | KC851842 | 12RS1402-13/APMV1/CK/ Ethiopia |
|
Fenti et al 2013 |
II | KC851853 | 12RS1402-83/APMV1/CK/ Ethiopia |
|
Aldous et al .2003 |
V | AY 175654 |
AV 1300/95 MG-10-05-CTanzania |
|
Aldous et al .2003 |
V | AY 175656 |
AV 1300/95 TB-02-24-DTanzania |
|
Aldous et al .2003 |
V | AY 175728 |
AV 1300/95 MG-04-08CTanzania |
|
Da Silva et al 2020 |
V | Mt33577389261 |
Chicken/Tanzania /2016 |
| Da Silva et al 2020 | V | Mt335727 | Chicken/ Tanzania /2011 |
|
Snoeck et al ;2009 |
VI | AJ880277 | Pigeon paramyxovirus-1/IT-227/82 |
|
Aldous et al .2003 |
VI | AY 175720 | Panvac (2/P2 UQ vet path AuEthiopia |
|
Snoeck et al ;2013 |
VIg | JQ039395 | pigeon/Nigeria/VRD07-173/2007 |
|
Snoeck et al ;2013 |
VIg | JQ039385 | dove/Nigeria/VRD07-163/2007 |
|
Snoeck et al ;2013 |
VII | KC205475 | chicken/Ethiopia/Dhera/2011 |
|
Snoeck et al ;2013 |
VIb | AY288997 | chicken/Kenya/139/90 |
|
Snoeck et al ;2013 |
VIh | JQ039391 | pigeon/Nigeria/VRD07-231/2007 |
|
Snoeck et al ;2013 |
VIh | JQ039387 | pigeon/Nigeria/VRD08-37BRpe(7-9)/2008 |
|
Snoeck et al ;2013 |
VIa | EF030963 | chicken/Cameroon/CAE11-855/2011 |
|
Snoeck et al ;2013 |
VIa | EF030951 | pigeon/South Africa/PIZA05N277/2005 |
|
Snoeck et al ;2013 |
VIa | EF030953 | chicken/South Africa/CKZA06N606/2006 |
|
Snoeck et al ;2013 |
VIa | AY445669 | dove/South Africa/DOZA05N240/2005 |
|
Snoeck et al ;2013 |
VIa | EF030958 | chicken/South Africa/469/2002 |
|
Snoeck et al ;2013 |
VIa | EF030962 | dove/South Africa/DOZA06N589/2006 |
|
Snoeck et al ;2013 |
VIa | EF030950 | pigeon/South Africa/PIZA04N230/2004 |
|
Snoeck et al ;2009 |
VIa | EF030957 | pigeon/South Africa/PIZA06N642/2006 |
|
Snoeck et al ;2009 |
VIb | AY288997 | dove/South Africa/DOZA06N549/2006 |
|
Chake et al 2013 |
VIf | Kc205476 | NDV/ Ethiopia/ 2011 |
|
Chake et al 2013 |
VIf | Kc205475 | NDV/ Ethiopia/ 2011 |
|
Megahed et al 2018 |
VII | KX231853 |
NDV/Chicken/Egypt/2/2015 |
| Megahed et al 2018 | VII | KX231854 |
NDV/Chicken/Egypt/4/2015 |
| Megahed et al 2018 | VII | KX231852 |
NDV/Chicken/Egypt/1/2015 |
| Megahed et al 2018 | VII | KX231851 |
NDV/Chicken/Egypt/3/2015 |
| Fenti et al 2013 | VII | KC851841 | 12RS1402-39/APMV1/CK/ Ethiopia |
|
Fenti et al 2013 |
VII | KC851848 | 12RS1402-67/APMV1/CK/ Ethiopia |
|
Radwan et al ,2013 |
VIId | JX173098 | NDV/Chicken/Giza/Egypt/MR0/2012 |
|
Abolink et al.,2017 |
VIIh | HQ697256 | Chicken/Makassar/003/09 |
|
Abolink et al.,2017 |
VIIh | HQ697255 | Chicken/Sukorejo/019/10 |
|
Abolink et al.,2017 |
VIIh | KR074405 | IBS005/11 |
|
Abolink et al.,2017 |
VIIh | KX231366 | NDV/chicken/ Mozambique/1205/2011 |
|
Abolink et al.,2017 |
VIIh | KT760568 | Chicken/Guizhou/1032/ 2012 |
|
Abolink et al.,2017 |
VIIh | KU523524 | NDV/chicken/ Mozambique/466/2012 |
|
Abolink et al.,2017 |
VIIh | JX870619 | KH-cmb08-2/12 |
|
Abolink et al.,2017 |
VIIh | KU523528 | NDV/chicken/ Mozambique/658/2012 |
|
Abolink et al.,2017 |
VIIh | KU523529 | NDV/chicken/ Mozambique/584/2013a |
|
Abolink et al.,2017 |
VIIh | KR815908 | Turkey/South Africa/ N2057/2013a |
|
Abolink et al.,2017 |
VIIh | MF622045 | Chicken/South Africa/ RBNW-1/2013a |
|
Abolink et al.,2017 |
VIIh | MF622047 | Chicken/South Africa/ RBNW-3/2013a |
|
Abolink et al.,2017 |
VIIh | MF622037 | Chicken/South Africa/ 239391/2013a |
|
Abolink et al.,2017 |
VIIh | KU523533 | NDV/chicken/ Mozambique/622/2014 |
|
Abolink et al.,2017 |
VIIh | MF622034 | Chicken/South Africa/ 32995/2015a |
|
Abolink et al.,2017 |
VIIh | MF622044 | Chicken/South Africa/ N2683/2015a |
|
Abolink et al.,2017 |
VIIh | MF622042 | Chicken/Zambia/ Mbeweka/2015 |
|
Abolink et al.,2017 |
VIIh | KX231368 | NDV/chicken/ Mozambique/192A/2016 |
|
Hertzeg et al. (1998 |
VIIb | AF136767 | ZA 20/93/ South Africa |
|
Hertzeg et al. (1998 |
VIIb | AF136768 | ZA 25/93/ South Africa |
|
Hertzeg et al. (1998 |
VIIb | AF136769 | ZA 26/93/ South Africa |
|
Hertzeg et al. (1998 |
VIIb | AF136772 | ZA 33/94/ South Africa |
|
Hertzeg et al. (1998 |
VIIb | AF136774 | ZA 35/95/ South Africa |
|
Hertzeg et al. (1998 |
VIIb | AF109876 | ZA 360/95/ South Africa |
|
Hertzeg et al. (1998 |
VIIb | AF136775 |
MZ 13/94 Mozambique |
|
Hertzeg et al. (1998 |
VIIb | AF136776 |
MZ 35/94 Mozambique |
|
Aldous et al .2003 |
VIIb | AY 175640 | AV 990/99 -1145 South Africa |
|
Aldous et al .2003 |
VIIb | AY 175650 | AV 1111/92 X 988Sweden |
|
Aldous et al .2003 |
VIId | Y 175641 | AV 990/99 96-0842South Africa |
|
Ahmed et al 2011 |
VIId |
(MF418017 |
(NDV/Chicken/EG-MN/NRC/2015 |
|
Ahmed et al 2011 |
VIId | MF418018 | NDV/Chicken/EGQU/NRC/2015 |
|
Ahmed et al 2011 |
VIId | MF418019 | NDV/Chicken/EG-SH/NRC/2015 |
|
Ahmed et al 2011 |
VIId |
(MF418020 |
NDV/Chicken/EG -SH2/ NR/2015 |
|
Orabi et al 2016 |
VIId | KY075880 | NDV/chicken/Egypt/Sharkia7/2016 |
|
Orabi et al 2016 |
VIId | KY075882 | NDV/chicken/Egypt/Damietta9/2016 |
|
Hertzeg et al. (1998 |
VIII | AF136764 | ZA 16/90/ South Africa |
|
Hertzeg et al. (1998 |
VIII | AF136765 | ZA 17/90/ South Africa |
|
Hertzeg et al. (1998 |
VIII | AF136766 | ZA 18/90/ South Africa |
|
Hertzeg et al. (1998 |
VIII | AF136773 | ZA34/94/ South Africa |
|
Hertzeg et al. (1998 |
VIII | AF136762 | ZA 5/68/ South Africa |
|
Da Silva et al 2020 |
XIII | Mt335739 | Chicken/ Tanzania /2016 |
|
Da Silva et al 2020 |
XIII | Mt335748 | Chicken/ Tanzania /2017 |
|
Snoeck et al ;2013 |
XIVa | JQ039386 | chicken/Nigeria/VRD08-36/2008 |
|
Snoeck et al ;2013 |
XIVa | JN872165 | Chicken/Niger/VIR 1377-7/2006 |
|
Snoeck et al ;2013 |
XIVa | FJ772452 | chicken/Niger/1377-8/2006 |
|
Snoeck et al ;2013 |
XIVb | JQ039390 | chicken/Nigeria/VRD07-233/2007 |
|
Snoeck et al ;2013 |
XIVb | JX546245 | chicken/Benin/463MT/2009 |
|
Snoeck et al ;2013 |
XIVb | JF966386 | chicken/Mali/ML029_07/2007 |
|
Snoeck et al ;2013 |
XVIIb | FJ772446 | avian/Nigeria/913-1/2006 |
|
Snoeck et al ;2013 |
XVIIa | JX546243 | chicken/Benin/373GC/2009 |
|
Snoeck et al ;2013 |
XVIIa | JX546244 | chicken/Benin/376GT/2009 |
|
Snoeck et al ;2013 |
XVIIa | FJ772486 | avian/Nigeria/3724-6/2008 |
|
Snoeck et al ;2013 |
XVIIa | FJ772469 | chicken/Niger/2602-348/2008 |
|
Snoeck et al ;2013 |
XVIIa | FJ772472 | chicken/Niger/2602-468/2008 |
|
Snoeck et al ;2013 |
XVIIa | JF966385 | chicken/Mali/ML007_08/2008 |
|
Snoeck et al ;2013 |
XVIIa | FJ772463 | chicken/Burkina Faso/2415-580/2008 |
|
Snoeck et al ;2013 |
XVIIa | FJ772458 | chicken/Burkina Faso/2415-361/2008 |
|
Snoeck et al ;2013 |
XVIIa | JX546247 | chicken/Benin/488MT/2009 |
|
Snoeck et al ;2013 |
XVIIa | FJ772449 | avian/Nigeria/913-33/2006 |
|
Snoeck et al ;2013 |
XVIIa | JQ039392 | avian/Nigeria/VRD07-733/2007 |
|
Snoeck et al ;2013 |
XVIIa | JQ039394 | chicken/Nigeria/VRD07-410/2007 |
|
Snoeck et al ;2013 |
XVIIa | FJ772478 | chicken/Cameroon/3490-149/2008 |
|
Snoeck et al ;2013 |
XVIIa | FJ772484 | chicken/Cameroon/3490-147/2008 |
|
Snoeck et al ;2013 |
XVIIa | JQ039393 | chicken/Nigeria/VRD07-141/2007 |
|
Snoeck et al ;2013 |
XVIIa | FJ772475 | chicken/Niger/2602-605/2008 |
|
Snoeck et al ;2013 |
XVIIa | FJ772481 | chicken/Niger/2602-625/2008 |
|
Da Silva et al 2020 |
XVIII | Mt335753 |
2018Chicken/Ghana/ |
|
Snoeck et al ;2013 |
XVIIIa | JF966388 | chicken/Mali/ML225_08/2008 |
|
Snoeck et al ;2013 |
XVIIIa | JF966389 | guinea fowl/Mali/ML038_07/2007 |
|
Snoeck et al ;2013 |
XVIIIb | JN942101 | Finch/Eastern Hemisphere/1409-12/2008 |
|
Snoeck et al ;2013 |
XVIIIb | FJ772466 | chicken/Ivory Coast/2601/2008 |
|
Snoeck et al ;2013 |
XVIIIb | JX390609 | chicken/Togo/AKO18/2009 |
|
Snoeck et al ;2013 |
XVIIIa | JN872157 | chicken/Mali/ML008_09/2009 |
|
Snoeck et al ;2013 |
XVIIIa | JN872157 | GWH/Eastern Hemisphere/5801-22/10 |
|
Snoeck et al ;2013 |
XVIIIa | FJ772455 | avian/Mauritania/1532-14/2006 |
|
Snoeck et al ;2013 |
XVIIIb | HF969127 | chicken/Ivory Coast/CIV08-069/2007 |
|
Snoeck et al ;2013 |
XVIIIb | HF969126 | duck/Ivory Coast/CIV08-062/2006 |
|
Snoeck et al ;2013 |
XVIIIb | JX390609 | chicken/Togo/AKO18/2009 |
|
Snoeck et al ;2013 |
XVIIIb | HF969216 | chicken/Nigeria/NIE11-1286/2011 |
|
Snoeck et al ;2013 |
XVIIIa | HF969179 | chicken Ivory Coast/CIV08-026/2007 |
|
Snoeck et al ;2013 |
XVIIIa | FJ772455 |
avian/Mauritania/1532-14/2006 |
in neighboring territories, Nigeria applied the prophylactic vaccination against NDV, using the Komarov strain of Newcastle disease vaccine obtained from the Onderstepoort Laboratories, Pretoria, South Africa. However, they confirmed the first outbreak of NDV in Nigeria at Ibadan province during December 1952, which they confirmed by laboratory diagnosis in February 1953 (Hill et al., 1953).
The first documented evidence of ND in Chad occurred in 1954 since then, it occurs so epizootic and endemic (Provost et al., 1968). The first documented evidence of ND in Uganda occurred in 1955 (Mukabii, 1992). In July 1960, for the first time they diagnosed mild lento genic type of NDV in South Africa, known as the ‘American type’, was. The strain of the virus induced mild clinical signs that the careful observer can see only (Kluge, 1964). Up to our knowledge, the first documented evidence of NDV in Ethiopia dates back to 1978 when an outbreak occurred in Eritrea then spread to the Northern part of the country. Epidemiological and virological studies on intensively reared commercial chicken revealed that the velogenic strains are widely distributed through Ethiopia (Nasser, 1998). In the Sahara desert region, they characterized virulent isolates NDV of in Morocco during 1982 (Bell, 1985).
Nowadays, all African countries reported ND as being the most serious problem in live backyard rural poultry (Bell, 1991, 1992). Hence, it has been reported in the traditionally managed village poultry from Morocco (Bell and Moulodi, 1988), Mauritania (Bell et al., 1990a), Ethiopia (Bawke et al., 1991), Nigeria (Olabode et al., 1991), Sudan (Fadol, 1991), the Ivory Coast (Couacy-Hymann et al., 1991), Mozambique (Fringe and Dias, 1991) and Uganda (George, 1991).
Genotypes of NDV in Africa
The NDV phylogenetic analysis is an effective technique for evaluating the epidemiological relationships among the NDV isolates present in different parts of the world (Mase et al., 2002).
Currently two nomenclatures systems are in use for the classification of NDV, based on the phylogenetic analysis, a system that divides NDV into six lineages with several sub-lineages. As example, lineages 3 and 4 were further classified into four sub-lineages (a- d) whereas lineage 5 was divided into five sub-lineages (a -e). This system had several limitations as they focused on the restriction enzyme analysis and partial sequence of the F-gene (Ballagi-Pordany et al., 1996; Aldous et al., 2003).
The used second system based on full F or full genome sequences (Czegledi et al., 2006). It therefore categorized NDV into two main classes; I- II into one serotype, in which class II further split up to 18 genotypes they (Diel et al., 2012; Courtney et al., 2013; Snoeck et al., 2013). The comparison between the two systems revealed that sub-lineages 3a, 3b, 3c and 3d correlated to genotypes III, IV,V and VIII and sub-lineages 5a to 5d correlated to genotypes VII a to d (Aldous et al., 2003). However, novel subgenotypes VIIk are recently characterized (Molini et al., 2017).
Recurrent outbreaks of NDV attack the different regions of the Africa continent for years, despite of the previous the information about the genetic characteristics of the virulent strains circulating in that region is still limited. Until now, twelve NDV genotypes, I, II, IV, V, VI, VII, VIII, XI, XIII, XIV, XVII and XVIII, isolated from various African countries in different periods (Da Silva et al., 2020).
Genotype II
It belongs to class II NDVs and is a mixture of lentogenic and velogenic strains (Kim et al., 2008), which were firstly identified in North America. Genotype II virulent, Beaudette C/1945 and Texas/GB/1948, and lentogenic, Hitchner/B1/47 and LaSota/1946, strains were isolated during 1940 in the United States. The lentogenic strains commercialized and they used as vaccines in poultry (Ballagi-Pordany et al., 1996; Millar et al., 2010).
They isolated NDV strains from South Africa in a period from 1999 identified as genotype II lentogenic strains and suggested that its source the commercial vaccine. Thus, no true lentogenic wild type NDV strains they identified in this study (Abolink et al., 2004). A few years later, molecular characterization of 11 NDV strains isolated from Egypt during the period 1996 -2005 revealed that they belong to genotype II and VI, suggesting that these genotypes are the most abundant NDV genotypes circulating in the Egyptian poultry flocks (Saad et al., 2010). In Egypt, sequences analysis of the F genes from virulent NDV 2006 Egyptian isolates revealed that they belong to genotype II. Further, they are closely related to other strains isolated in Egypt in 2005 (Mohamed et al., 2009) and the North American strain, which strength the suggestion that those strains are probably the most abundant genotype in the bird population in Egypt (Mohamed et al., 2011). Furthermore, NDV strains of genotype II which were similar or variant from LaSota strain were identified in Nigeria and Burkina Faso in 2006 /2007 (Snoeck et al., 2009) an in northwest Ethiopia (Fentie et al., 2013). In northwest Ethiopia, recent study concluded that live chicken market plays important role in spreading and dissemination of NDV in chicken. They isolated NDV from apparently healthy appearing birds in all seasons of the year with high percent in pre-rainy dry season, showing evidence for climatic and socioeconomic aspects as risk factor in the occurrence of ND in chicken (Haile and Fentie, 2020).
Genotype V
Genotype V had started to emerge in the Americas for the first time around the 1970s and disseminate to the European continent in 1980s, scientists have recently reported its introduction to Africa in Eastern region (Aldous et al., 2003; Yongolo et al., 2011). The previous suggesting the increase of the genotype geographic distribution (Sabra et al., 2017). It possesses mainly pathogenic strains which sub classified in to several genotypes (a- d). In Tanzania, the analysis of NDV strains isolated between 1994 and 1995 revealed that they belong to genotype V and it may spread through migration of wild birds or live birds markets (Yongolo et al., 2011). Consistently, healthy live birds in live poultry markets act as reservoir for virulent NDV (Byarugaba et al., 2014). A recent study suggested possible cross border spread of velogenic NDV between Kenya and neighboring Uganda through live bird trade (Ogali et al., 2018). The Ugandan NDV strains has low genetic diversity suggesting low evolution rate with amino acid mutations in the HN protein that differ from the Kenyan strains, suggesting independent evolution. The first complete genome sequence analysis of subgenotype Vd was performed on single NDV strain isolated also from live poultry market in the Mbeya region, Tanzania (Goraichuk et al., 2019). Later on, similar study reports the first complete genome sequence of NDV from backyard chicken in Kenya. The three isolates characterized to be velogenic by MDT and ICPI. Those were phylogenetically related to genotype V strains, and form a distinct cluster together with NDV strains from the East African countries of Uganda and Tanzania to form the newly characterized subgenotype Vd (Ogali et al., 2020). Another recent study utilized the third-generation portable sequencing device in identification of nine Tanzanian NDV strains belonging to genotype V. The NDV strains of this study were collected from Morogoo, Tanzania in a period between 2011and 2017 and were closely related to other isolates from Mbeya, Kenya and Uganda (Da Silva et al., 2020). Circulation of closely related NDV strains of genotype V appeared within these three East African countries and the unrestricted movement of cross-border trade of live birds might be the main reason for endemic situation of this genotype in this region (Byarugaba et al., 2014; Msoffe et al., 2019).
Genotype VI
Genotype VI viruses, pigeon paramyxoviruses 1, is usually isolated from Columbidae and sub classified in to subgenotypes VIa to k. Isolates of NDV belonging to the genotype VIb were mainly restricted to the western African region. In 2007, two sequences of genotype VI with two nucleotides mismatching obtained from a parrot and pigeon in Nigeria (Snoeck et al., 2009). Further investigation confirmed the circulation of genotype VIb in pigeons and rural poultry samples from six Nigerian states during 2007-2008 (Vam Borm et al., 2012). Introduction of putative subgenotypes derived from genotype VI, VIh and VIi, was reported in 2013 from pigeon samples (Snoeck, 2013). The existence of genotype VIh from Pigeon samples and their clustering with foreign strains such as Pigeon/Argentina/Capital_3/97 indicated strong evidence for the responsibility of the wild birds in the introduction of genotype VI to the Western region of the African continents. The Nigerian subgenotype VIg isolates were highly correlated to other NDV strains from Egypt, Russia and Ukraine while the subgenotype VIh isolates have high homology with wild bird isolates from Kenya. Further, the Nigerian subgenotypes VIi were closely related with Italian strains from outbreak in doves. These close genetic relationships among the isolates could be of epidemiological significance and strongly recommend a previous common origin during their natural selection (Snoeck et al., 2013).
Not only restricted to the western part of the African continent, as the free birds-specificity of genotype VI, led to dissemination of the genotype to other countries such as Egypt (VIg) and central Ethiopia (VIf) (Sabra et al., 2017; Chaka et al., 2013).
Genotype VII
It represents NDV strains isolated during the epizootics of Mozambique and South Africa (1990-1995), hence they classified as subgenotype VIIb (Herczeg et al., 1999). The authors suggested that the genotype might be emerged to the country from the Far East and western European countries.
Subgenotype VIId was initially isolated during 1999 from shanghai goose flocks which were suffered from moderate mortality rates in goslings. The causative virus strain, SF02, was initially termed as Goose paramyxovirus thereafter it was identified as genotype VIId. NDV strains belong to subgenotype VIId are highly pathogenic to different domesticated birds with intra-cerebral pathogenicity indexes (ICPIs) ranged from 1.80 to 1.94 (Jinding et al., 2005; Liu et al., 2003; Zou et al., 2002; Zou and Gong 2003). The phylogenetic analysis of NDV strains isolated from Egypt between 2011 and 2012 revealed that they belong to genotypeVII subgenotype d and closely related to the Middle East isolates (Radwan et al., 2013). Since 2011, NDV outbreaks caused by genotype VII occur in several vaccinated and non-vaccinated poultry in different governorates of Egypt, there is a strong probability that the Egyptian VIId strains are introduced from China or Middle Eastern countries such as Israel and enter Egypt through infected poultry products or wild birds (Hussein et al., 2014; Megahed et al., 2018).
Two virulent NDV isolates was clustered with genotype VIId (lineage 5) and were similar to those NDV isolates from northeastern African countries (Sudan and Egypt), which were reported to GenBank between 2004 and 2012. The close genetic similarity of the northern Ethiopian virulent isolates with that of the Sudanese and Egyptian isolates provides evidence for potential epidemiologic links between outbreaks of NDV in these countries, likely due to the geographical proximity and movement of poultry among borders (Fentie et al., 2013). Recent study includes molecular and phylogenetic characterization of 116 tissue / organ collected from NDV outbreak in Egypt and reveals that they are velogenic strains with high identity to velogenic strains from Jordan, Israel and Turkey. Phylogenetic analysis showed that they belong to genotypes (VIId,VIIa and II) and VIId is predominant genotype circulating in Egypt (Mouhamed et al., 2020).
In the period between 2002 and 2007, different virulent NDV strains isolated from backyard farms and live bird markets in four Sub-Saharan countries in the West Africa, Nigeria, Niger, Burkina Faso and Cameroon. The strains were clustered within subgenotype VIIf and g which represent NDV variants indigenous to West Africa (Snoeck et al., 2009).
Subgenotype VIIh has introduced to South-East Asia and caused serious outbreaks for the poultry flocks in Malaysia, Indonesia, southern China, Vietnam and Cambodia. It was recorded in the African continent in 2011 during outbreaks in poultry flocks. Thereafter, the virus spread north through Mozambique (Mapaco et al., 2016) into the north-eastern provinces of Zimbabwe and then south towards Botswana and South Africa causing serious outbreak in 2013 (Abolnik et al., 2017). Further, the virus transmitted from Mozambique toward neighboring countries such as Malawi and Zambia. Recent study determines the NDV genotype circulating in Botswana. Phylogenetic analysis of fourteen samples that collected from NDV outbreak in 2014, 2018 and 2019 reveals that they clustered in genotype VII. They closely related to viruses from South Africa and Mozambique than other southern African countries (Nambia, Zambia and Zimbabwe) (Kgotlele et al., 2020). In Fact, Abolnik and his colleagues suggested that the absence of genotype VIIh outside of South-East Asia and Southern Africa is strong evidence that wild migratory birds are not responsible for disease dissemination. Hence, there are strong evidences that illegal live bird trade or infected wastes from ships are the source of the virus emergence to Mozambique in 2011.
Genotype VIII
Genotype VIII NDV strains are geographically restricted viruses which were originated from Southeast Asia between 1979 and 1985 (Aldous et al., 2003; Liang et al., 2002), this genotype has emerged in South Africa since decades (Herczeg et al., 1999). Hence, it was characterized in South African strains that were detected in the 1960 and in the period from 1990 to 1995. However, no record for the isolation of this genotype has been available since June 2000 (Abolnik, 2007).
Genotype XI
NDV strains belong to genotype XI are mainly virulent. It is geographically restricted in Africa. Hence, reporting only in Madagascar, where it circulated between the wild birds and domestic chicken (De Almeida et al., 2013).
Genotype XIII
Virulent genotype, which subclassified in to three subgenotypes, namely, XIIIa, XIIIb, and XIIIc. Subgenotype XIIIa strains have been isolated during poultry outbreaks in Europe, Africa, and the Middle East, but subgenotypes XIIIb and XIIIc remain localized to India and Pakistan (Dimitrov et al., 2016). In Africa, genotype XIII strains have been recovered from an ostrich in South Africa (1995), chickens in Burundi (2008), and passerines in Tanzania (2010) (Cattoli et al., 2010; Benson et al., 2015). Recently, this genotype has been isolated from Zambia, Burundi, South Africa and Tanzania (Msoffe et al., 2019; Da Silva et al., 2020).
Genotypes XIV, XVII and XVIII
These moderately recent discovered regional genotypes found endogenous in West and Central Africa.
Genotypes XIV: It is NDV variant that endogenous in West African countries such as Nigeria, Niger, Cameroon and Burkina Faso. It was initially isolated from live poultry market and backyard flocks in the period from 2002 to 2008 and was initially termed as lineage 5f (Snoeck et al., 2008; Solomon et al., 2012) or 7d (Cattoli et al., 2010). Thereafter it was renamed as genotype XIV according to the new classification (Snoeck et al., 2013) with two subgenotypes, a and b or 1 and 2 (Dimitrov et al., 2019). Fourteen and 33 strains of subgenotype XIVa and XIVb, respectively, were detected in different Nigerian cities in the period from 2007 to 2011 (Snoeck et al., 2013). Interestingly, all the sequenced strains possess the cleavage site specific for NDV virulent pathotype. Accordingly, pathogenicity study trial revealed that genotype XIV isolated from West Africa is velogenic viscerotropic pathotype that caused severe clinical signs or mortality by the day 4-post infection (Susta et al., 2014). Further, it led to systemic infection and necrosis in the lymphoid tissue and gastrointestinal tract. The first complete genomic sequence of genotype XIVb NDV strain (duck/Nigeria/NG-695/KG.LOM.11-16/2009) that isolated from healthy duck in live poultry market in Nigeria was reported in (Shittu et al., 2016).
Genotype XVII: Snoeck and his colleagues defined it together with genotype XVIII (Snoeck et al., 2013) according to criteria described by (Diel et al., 2012). They isolated it from domestic birds from different countries in West Africa plus Central African Republic (Snoeck et al., 2013) that may suggest the genotype ability to spread across the continent. The virus classified into two subgenotypes with virulent characteristics.
Genotype XVIII: Viruses of class II genotype XVIII, previously identified as lineage 7a (Cattoli et al., 2010) with a high correlation with genotypes XIV and XVII. In fact, both genotypes XVII and XVIII have been suggested to be the same genotype (Desingu et al., 2016). However, this conclusion was not accepted by Snoeck and Muller (2016) as they stated that Desingu and colleagues used incorrect bioinformatics parameters to analyze NDV genotyping. However, until now it has still been classified as distinctive genotype divided in to two subgenotypes XVIIIa and XVIIIb by (Snoeck et al., 2013) or the more updated XVIII.1 and XVIII.2 by (Dimitrov et al., 2019). West African countries seem to have the most complex situation in terms of genetic diversity of NDV genotypes and subgenotypes. The two subgenotypes, XVIII.1, and XVIII.2 have been found independently circulated or either co-circulated together or with other genotypes, such as subgenotype XVIIa, in Côte d’Ivoire, Mauritania, Mali, Togo and Nigeria since 2006 or may be earlier (Cattoli et al., 2010; Snoeck et al., 2013). In 2018, four sequences obtained from two distant cities in Ghana, Wa and Pokoasi, found to be belong to subgenotype XVIII.2 and were highly correlated to an isolate from Mali (JX518886) (Da Silva et al., 2020). With the fact that it is geographically restricted or regional genotype in the Western Africa, the international trade of exotic birds can play a role in its worldwide dissemination.
NDV Challenges in Africa
Based on the available information, it is obvious that there is broaden ecological distribution of various NDV genotype across the continent. The continuous genetic modification in NDV with mutation in the cleavage site and the neutralizing epitopes could lead to what is called vaccination failure phenomena. Accordingly, all the previous together with the existence of wild birds and the unrestricted movement between neighbor countries border will obscure the problem of recurrent NDV outbreak in Africa.
Conclusion
ND is considering a major threat for poultry production in Africa. Several genotypes are isolated and their distribution in most of African countries was recorded. Genotype VII which is responsible for the fourth panzootic ND outbreak is also found in the region. Genetic diversity between NDV strains continue to grow mainly in central and West Africa where the recent discovered genotypes XIV, XVII, XVIII are geographically restricted. Africa is considering a reservoir for new virulent strains, the efficacy of the classical vaccinal strains against these recent strains need to be further studied. Wild, exotic birds, live bird market and illegal trading play important role in entrance and spreading of NDV strains and their control is necessary.
acknowledgements
We are thankful to the scientific committee of The 15th Scientific Conference of Veterinary Medicine entitled human and animal Health and safety: Reality and Ambition.
Conflict of Interest
There is no conflict of interest.
authors contribution
Wala Mohamed and Ola Hassanin collected and analyzed the previously published scientific articles as well as designed and wrote the first draft of the review. Mohamed Megahed reviewed and approved the final version of the review.
References