Journal of Animal Health and Production
Research Article
Differential Methylation of Melanin-related Epigenetic Genes during Brindle Cattle Growth
Kyoung-Sub Jung1, Sang-Hwan Kim2, Jong-Taek Yoon2,3*
1Major in the Animal Biotechnology, Graduate School of Future Convergence Technology, Hankyong National University, Anseong, Gyeonggi-do, 456-749, Korea; 2Institute of Genetic Engineering, Hankyong National University, 327, Jungang-ro, Ansung, Gyeonggi-do, 17579, Korea; 3Department of Animal Life and Environment Science, Hankyong National University, 327, Jungang-ro, Ansung, Gyeonggi-do, 17579, Korea.
Kyoung-Sub Jung and Sang-Hwan Kim contributed equally to this work.
Abstract | Coat pigmentation in mammals is the determined by the relative distribution of pheomelanin and eumelanin. The phenotype of these two pigments, when expressed in melanocytes, is regulated by the mechanism of melanogenesis and the agouti locus alleles. Therefore, the present study analyzed mutations in the coat color-related genes in two Korean cattle breeds i.e., Hanwoo and Korean brindle cattle. According to results the coat color-related epigenetic markers [differentially methylated CpGs (DM CpGs)], by reduced representation bisulfite sequencing showed that brindle cattle has the biological functions of DM CpG-associated genes. In particular, the present study analyzed the genetic variation of the post-forming factor and expression patterns of the genes that affect the coat color of brindle cattle and revealed that the determination of coat color of brindle cattle was distinct from that of other Korean Hanwoo. Thus, the mutation or activity of major factors of MAPK were involved in melanogenesis by the activation of melanocortin 1 receptor (MC1R). In addition, the MARK mechanism significantly affected melanogenesis, even at lower concentrations of MC1R. These results suggested the possibility that hair color could be determined by the mutation of the agouti gene in the black coat color section. Therefore, the results of this study could help in the modulation of coat color and positive selection of the Korean brindle cattle.
Keywords | Brindle cattle, Epigenetic genes, Melanin, Melanocytes, Methylation
Received | March 08, 2020; Accepted | May 11, 2020; Published | May 28, 2020
*Correspondence | Jong-Taek Yoon, Institute of Genetic Engineering, Hankyong National University, 327, Jungang-ro, Ansung, Gyeonggi-do, 17579, Korea; Email: ohmyfamily@naver.com; jtyoon@hknu.ac.kr
Citation | Jung K-S, Kim S-H, Yoon J-T (2020). Differential methylation of melanin-related epigenetic genes during brindle cattle growth. J. Anim. Health Prod. 8(2): 80-88.
DOI | http://dx.doi.org/10.17582/journal.jahp/2020/8.2.80.88
ISSN | 2308-2801
Copyright © 2020 Popova et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Introduction
The history of the nation’s cattle began about 4,000 years ago with the hybridization of Indian zebu (Bos zebu) and European cattle (Bos taurus), and it has been reported that this hybrid was carried to Korea by the Chinese, Mongolians, and Manchurians. Kim et al. (2000) could not confirm the exact basis of the history and origin due to difficulties in securing samples. An important factor in determining the breed of livestock is the coat color. The change in the mechanism of melanogenesis, in turn, is very important in determining the coat color of mammals. Melanocortin 1 receptor (MC1R), which is activated by the introduction of melanocyte stimulating hormones (α-MSHs) in melanocytes, is a typical coat color determinant (Seo et al., 2007; Goud et al., 2019). It forms pheomelanin, which denatures red or yellow color through genetic mutation, and eumelanin, which is considered important in determining the coat color of mammals (Jackson, 1993; Robbins et al., 1993). It was reported that the offset action of the agouti factor in melanogenesis could interfere with the binding of α-MSHs to MC1R (Rieder et al., 2000; Choudhary et al., 2020), facilitating the production of pheomelanin. However, currently it is known that the expression of the agouti factor in the determination of coat color plays an important role in activating the expression or variation of MC1R (Girardot et al., 2005, 2006). Microphthalmia-associated transcription factor (MITF) activates transcription of the target genes of tyrosine (TYR), tyrosinase-related protein 1 (TYRP1), and dopachrome tautomerase (DCT) metabolism (Zhou et al., 2012; Shen et al., 2012). In other words, MITF is recognized as a higher gene in the control of the formation of complex melanosomes. MITF is produced and facilitated by Wnt signaling, which is known to play a crucial role in controlling cell fate, proliferation, and differentiation (Kang et al., 2011). However, according to Han et al. (2011), the genetic variation of the agouti gene is assumed to have no effect on the development of a Korean Hanwoo or hybrid offspring, and it does not play a role in coat color variation. Meanwhile, Brown (2012) reported that the typical black/brown coat color gradually changed to yellow in castrated bulls of South China. Lee et al. (2015) reported that E2 concentration in expressed yellow to brindle colors increased before the determination of the initial coat color, but gradually decreased after the brindle color was formed. Bennett et al. (2003) was involved in the study of the control of pigments by MSH and melanocortin receptor (MCR) and regulation of cortisol, which is related to the adrenocorticotropic hormone (ACTH) and MC1R. In addition, POH (pro-opiomelanocortin) derivatives MSH and ACTH were regulated by receptors (including cortisol) of pigments, hormones, and steroid hormones. Hirobe et al. (2011) reported that E2 injection in mice increased the coat pigmentation. Therefore, the present study analyzed the genetic variation of the post-forming factor and expression patterns of the genes that affect the coat color of brindle cattle and revealed that the determination of coat color of Korean brindle cattle was distinct from that of other Korean Hanwoo.
MATERIALS AND METHODS
Ethical approval
All animal procedures followed the protocol approved by the Animal Experimentation Ethics Committee at Hankyong National University (permission number: 2018-2).
Experimental animals
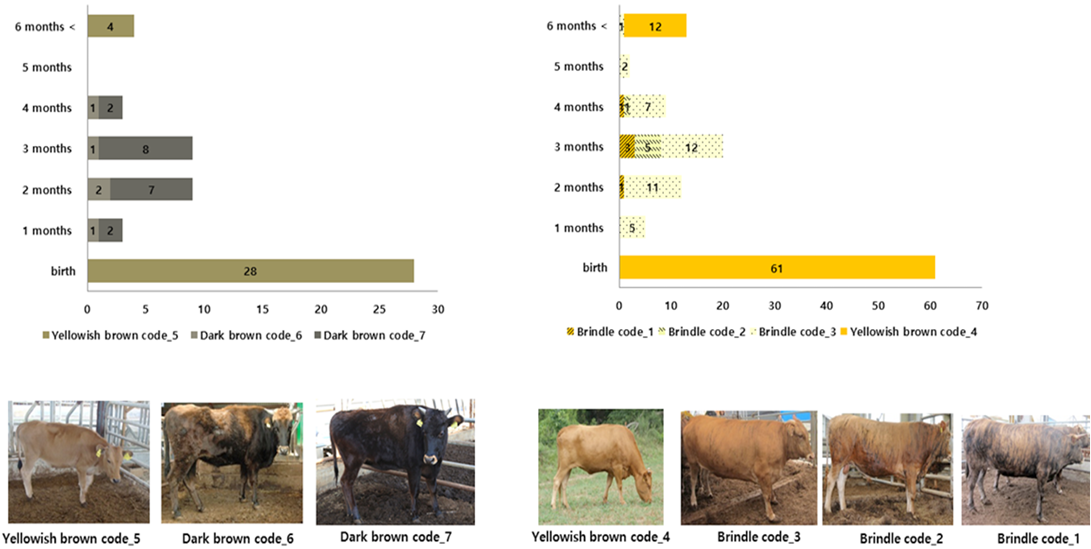
Brindle cattle used in this experiment were bred by the National Institute of Animal Health in Chungcheongbuk-do and were classified as brindle, black, and yellow (Figure 1), according to the classification method of Park et al. (2016). To analyze the patterns of epigenetic expression of coat color during different growth stages, DNA of young brindle cattle (who reached the age of six months), whose coat color did not change after birth and began changing at the age of less than one month, was extracted and used for analysis (Figure 2).
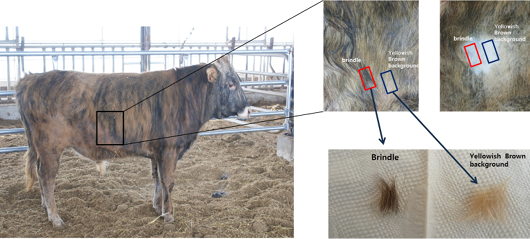
Figure 1: Samples for NGS analysis.
Next-generation sequencing (NGS)
The NGS analysis was conducted using Bovine genome assembly UMD 3.1. It included information about the unknown genes and variation in the genome sequences expressed on chromosomes, and it did not apply to the repetitive genome sequences on the same line. The sequence of bases passed through the standard Illumina Chastity filter was analyzed further, and the sequence analysis was adjusted to a 90 base pair (bp) reading to reduce errors in the sequence in the last reading section of the first reading. Short-read mapping was completed using BBWA ver. 0.5.9. Data that could not be read were removed after mapping. To analyze the single-nucleotide polymorphisms (SNPs) and insertions and deletions (InDels), SAM tools and additional filters were used as follows:
- 1. SNPs and InDels with overall quality less than 20 were removed.
- 2. Variants with too low or too high read depth were removed. First, we calculated the mean and standard deviation (SD) read depth for all variants. Then we set the minimum as 10% of the mean and maximum as mean read depth + 3*SD read depth.
- 3. Variants with less than one forward or reverse alternative allele were removed.
- 4. Variants within five bp of each other were removed.
- 5. SNPs within five bp of an InDel were removed.
- 6. InDels within 10 bp of each other were removed.
- 7. Variants that have sites that were not in the reference genome were removed.
Following the SNP and InDel analyses, NGS-SNP (Grant et al., 2011) was used to allocate functional classes to each SNP, and, where applicable, information on proteins and transcription proteins was provided. Functional analysis of the mechanism was performed using Ensembl release 68, Entrez Gene, NCBI, and UniProt (Flicek et al., 2011; Sayers et al., 2011).
Reduced representation bisulfite sequencing (RRBS) library preparation method
DNA (500 ng) was reacted with Msp1 (NEB) at 37°C for 24 h and then with Apek1 (NEB) at 75°C for 20 h. Digested DNA was refined using the MiniElute PCR Purification Kit (Qiagen). It was then end-repaired with T4 DNA polymerase (NEB), klenow fragment (NEB), T4 polynucleotide kinase (NEB), dNTP, and 1×polynucleotide kinase buffer. The response was seen at 20°C after 30 min. Blunt ended DNA fragment with klenow (3’-5’ exo-, NEB) was a-tailing at 37°C for 30 min. It was refined using the MiniElute PCR Purification Kit (Qiagen), and the adapter (illumina) was ligated using T4 DNA ligase (NEB) at 65°C for 15 min, after keeping it at 20°C for 15 min. DNA (160-420 bp) was selected using 2% agarose gel electrophoresis. According to the ZYMO EZ DNA manufacturer’s instructions, bisulfite was converted using the methylation-Gold Kit (ZYMO). The bisulfite-converted products were PCR-amplified using PfuTurbo Cxhotstart DNA polymerase (Agilent technology). Primer used was as follows: forward: 5’-AATGATACGGCGACCACCGAGAT-3’, reverse: 5’-CAAGCAGAAGACGGCATACGA-3’. The PCR conditions were 11 to 15 cycles at 94°C for 10 s, 58°C for 30 s, 72°C for 30 s, and 72°C for 5 min extension after 94°C for 1 min. The library was analyzed using the Agilent 2100 Bioanalyzer (Agilent Technologies) and quantified with real time PCR (RT-PCR).
Statistical analysis
RT-PCR results were analyzed for statistical significance using the SAS package (Statistical Analysis System, Institute, version 9.4, Cary, NC, USA). The data are shown as mean ± SD, and the significant difference among the groups was determined at p value <0.05.
Results
Patterns of genetic expression of black and yellow hair sections in brindle cattle
NGS analysis of specific and non-specific expression genes in the black and yellow hair sections of the brindle cattle (brindle color) showed that the genes expressed in these two sections formed about 18,726 of the EST gene from about 1,000 bp to 10,000 bp (Figure 3). Among the differently expressed groups in the GO term of the gene, three processes of fourteen biological processes, seventeen cellular components, and seven molecular functions showed significant differences. There was a significant difference (p <0.01) at the cellular component and biological process levels (cell cycle and activating factor). The results showed that the cell activating factors were different in the black and yellow sections of the brindle color. The number of genes expressed differently was high in the cellular component group in 321 biological processes, 702 cellular components, and 171 molecular functions, and a large number of melanosome intracellular non-membrane-bounded organelle related genes were found (Table 1). In this experiment, a comparison of the expression pattern of genes involved in melanogenesis, among the separated genes, confirmed that the expression of seven factors such as TYRP and MITF, which are related to melanocytes, in the black section was more than five times greater than that in the yellow zone. It was also confirmed that eumelanin of 49 major factors, such as MARK-related factors associated with melanogenesis, was involved (Figure 4). Thirty factors were found to be expressed more than five times in the black color section of brindle color, demonstrating that the mechanism and composition of melanogenesis in the yellow and black sections were different.
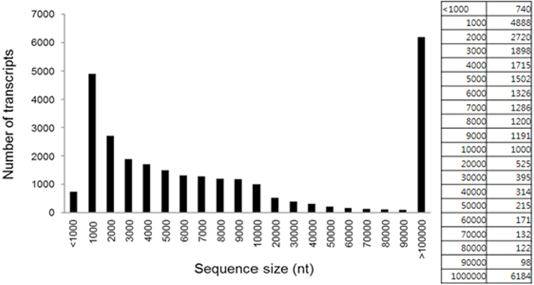
Figure 3: Length distribution and abundance of all transcripts identified in both the yellow background and brindle section.
The change in the coat color across growth stages
Analysis of the change in coat color at each growth stage predicted that 68% of 28 brindle cattle showed a change in coat color at about one to six months of age. Additionally, it was confirmed that 50% of 61 brindle cattle showed a change in coat color at about one to six months of age (Figure 5).
The change in DNA methylation across growth stages
Accordingly, the significant level of FDR <0.01 and methylation difference >25% was applied in the results of gene methylation analysis on DNA across growth stages of brindle cattle. About 1395 were selected for group comparisons (3 week vs. 6 month), and 1493 to 7551 DM CpGs were selected for individual comparisons.
Table 1: Top 30 highly expressed genes in the brindle section compared to the yellow background.
| GO terms | Total number of genes |
Template matching (P<0.01) |
||
| Biological process | GO:0007398 | ectoderm development | 17 | 6.05E-06 |
| GO:0008544 | epidermis development | 16 | 1.26E-05 | |
| GO:0060429 | epithelium development | 18 | 7.04E-04 | |
| GO:0060429 | epithelium development | 18 | 7.04E-04 | |
| GO:0030216 | keratinocyte differentiation | 7 | 7.63E-04 | |
| GO:0007264 | small GTPase mediated signal transduction | 28 | 0.001966 | |
| GO:0009913 | epidermal cell differentiation | 7 | 0.002496 | |
| GO:0018149 | peptide cross-linking | 7 | 0.002496 | |
| GO:0022610 | biological adhesion | 43 | 0.002652 | |
| GO:0007155 | cell adhesion | 43 | 0.002652 | |
| GO:0006468 | protein amino acid phosphorylation | 58 | 0.003019 | |
| GO:0030855 | epithelial cell differentiation | 11 | 0.004316 | |
| GO:0051056 | regulation of small GTPase mediated signal transduction | 26 | 0.004546 | |
| GO:0016337 | cell-cell adhesion | 22 | 0.007546 | |
| Cellular component | GO:0005856 | cytoskeleton | 98 | 1.66E-11 |
| GO:0045111 | intermediate filament cytoskeleton | 30 | 1.41E-10 | |
| GO:0005882 | intermediate filament | 30 | 1.41E-10 | |
| GO:0045095 | keratin filament | 21 | 1.44E-09 | |
| GO:0005911 | cell-cell junction | 27 | 3.65E-08 | |
| GO:0043296 | apical junction complex | 19 | 3.57E-07 | |
| GO:0016327 | apicolateral plasma membrane | 19 | 4.85E-07 | |
| GO:0044430 | cytoskeletal part | 64 | 1.44E-06 | |
| GO:0044430 | cytoskeletal part | 64 | 1.44E-06 | |
| GO:0030054 | cell junction | 38 | 6.75E-06 | |
| GO:0005923 | tight junction | 13 | 1.13E-04 | |
| GO:0070160 | occluding junction | 13 | 1.13E-04 | |
| GO:0001533 | cornified envelope | 5 | 7.05E-04 | |
| GO:0043232 | intracellular non-membrane-bounded organelle | 125 | 0.002658 | |
| GO:0043228 | non-membrane-bounded organelle | 125 | 0.002658 | |
| GO:0030057 | desmosome | 5 | 0.004801 | |
| GO:0005922 | connexon complex | 6 | 0.009066 | |
| Molecular function | GO:0016755 | transferase activity, transferring amino-acyl groups | 9 | 1.87E-05 |
| GO:0016755 | transferase activity, transferring amino-acyl groups | 9 | 1.87E-05 | |
| GO:0004713 | protein tyrosine kinase activity | 22 | 2.71E-04 | |
| GO:0005198 | structural molecule activity | 56 | 5.87E-04 | |
| GO:0004672 | protein kinase activity | 59 | 0.001415 | |
| GO:0003810 | protein-glutamine gamma-glutamyltransferase activity | 5 | 0.006306 | |
| GO:0004714 | transmembrane receptor protein tyrosine kinase activity | 11 | 0.007039 | |
On applying these DM CpG-associated genes, it was confirmed that the genes were expressed differently, and the brindle cattle (brindle color) identified the difference in the expression of many genes. In other words, 002072558324 objects, with the highest expression of brindle color, showed hypomethylation of 3,332 genes and hypermethylation of 4,219 genes after growth, while 002072550199 objects, which had the lowest expression of brindle color, showed hypomethylation of 1,460 genes and hypermethylation of 865 genes after growth (Table 2).
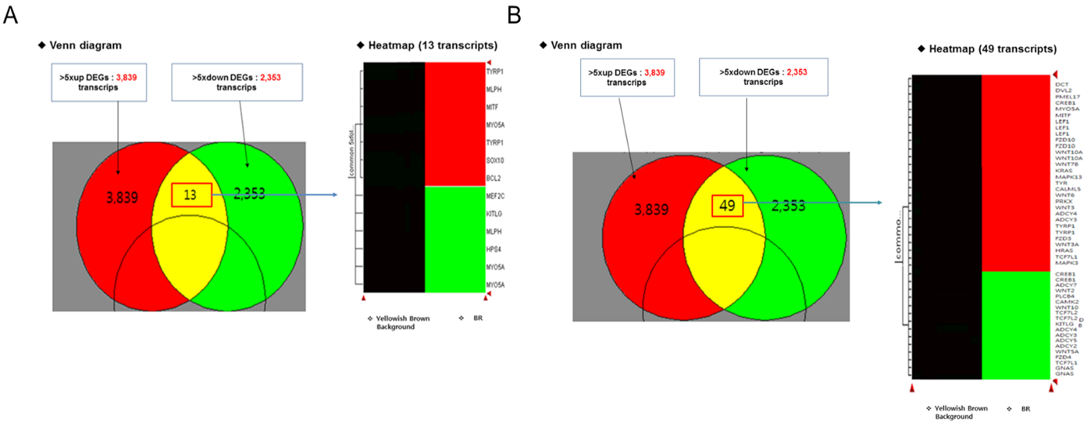
Figure 4: Profile analysis of the skin transcriptome in the brindle coat-Yellow zone: melanocyte-associated gene (common in >5×fold), A: melanocyte differentiation: 13 transcripts, B: melanogenesis differentiation: 49 transcripts.
Table 2: Differentially methylated CpGs with different coat color patterns.
| Sample |
# Differentially methylated CpGs |
|||
| Control | Case | All | Hyper | Hypo |
| 002072550199_160321_1 | 002072550199_160901_2 | 2325 | 865 |
1460* |
| 002072550211_160321_1 | 002072550211_160901_2 | 2359 |
1416* |
943 |
| 002072550318_160420_1 | 002072550318_160901_2 | 1493 | 773 | 720 |
| 002072550359_160420_1 | 002072550359_160901_2 |
3133* |
2576 | 557 |
| 002072558324_160317_1 | 002072558324_160902_2 |
7551** |
4219** |
3332** |
| brown | brindle | 1395 | 893 | 502 |
*,** Different letters within the same column represent a significant difference at P < 0.05 and P < 0.01 respectively.
Table 3: KEGG pathways for DM CpG-associated genes- brown vs. brindle (FDR <0.001, methylation difference >25%).
| Term | Count | % | PValue | Genes |
Fold |
FDR |
| bta04916: Melanogenesis | 24 | 1.355167 | 7.04E-06 | WNT5A, TYRP1, HRAS, GNAI2, MAP2K2, GNAI1, ADCY5, CREBBP, FZD1, KITLG, TCF7L2, WNT1, EDNRB, PLCB3, WNT3, PLCB4, CALM, WNT11, GNAS, CALML5, PLCB1, ASIP, CALM2, CALM1 | 2.828159 | 0.009283 |
| bta04310: Wnt signaling pathway | 25 | 1.411632 | 5.25E-04 | WNT5A, NKD1, CTBP2, CREBBP, FZD1, SMAD4, TCF7L2, WNT1, PLCB3, CCND1, SOST, PLCB4, WNT3, PSEN1, SFRP1, PPP3CC, WNT11, MAPK8, SOX17, BAMBI, PLCB1, AXIN2, MYC, CUL1, NFATC1 | 2.12886 | 0.690278 |
| bta04010: MAPK signaling pathway | 27 | 2.045455 | 0.009372 | PTPN7, FGFR1, HRAS, FGFR3, PDGFA, NF1, GNA12, MAP4K1, MAPK11, TAB1, TGFB2, PRKCB, RPS6KA5, RPS6KA6, DUSP2, DUSP1, MAPK3, MAPK8, PAK1, MAPK7, MAP3K14, GADD45B, MYC, MAP2K6, CACNA1B, NFATC1, FGF4 | 1.684266 | 11.67573 |
| Thyroid hormone signaling pathway | 15 | 1.136364 | 0.010108 | FXYD2, HRAS, PIK3CB, RXRA, CREBBP, RCAN1, PRKCB, HIF1A, DIO3, MAPK3, MED27, GATA4, RHEB, MYC, SLC9A1 | 2.128103 | 12.53706 |
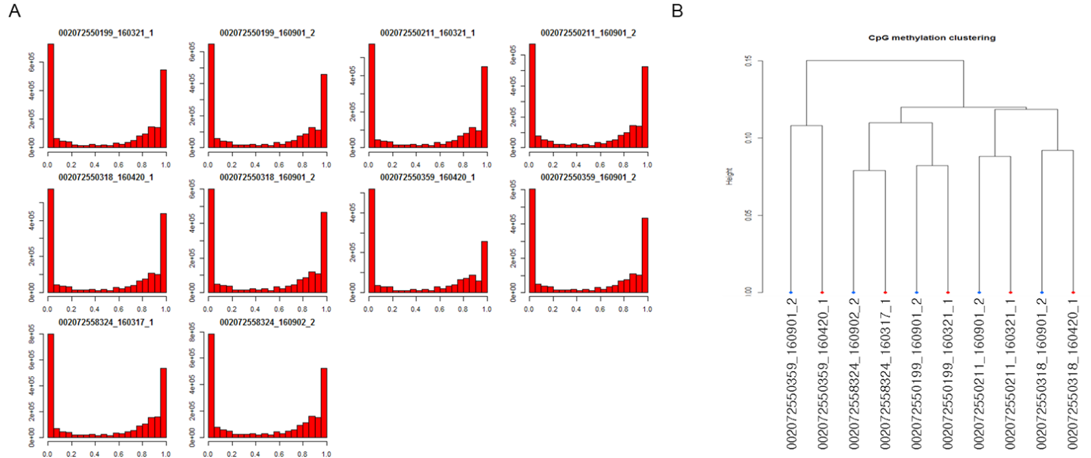
Figure 6: Methylation level distribution among brindle cattle- A: methylation level, B: clustering by CpG methylation.
The change in DNA methylation in individual growth stages
Hypomethylation in 201380 Section 1 of the CpG chromosome was done by 27.11% of 002007255019 objects, while hypomethylation in the 44256011 sections of the CpG chromosome 29 was done by 100% of 002072550211 objects at growth. Hypomethylation in 100178350 sections of the CpG chromosome 1 was done by 66.66% of 002072550318 objects, and hypermethylation in 48967783 sections of the CpG chromosome 11 was done by 62.5% of 002072550359 objects. Hypomethylation in 10195476 of the CpG chromosome 11 was done by 66.66% of 0020072550324 objects with distinctly expressed brindle color (Figures 6 and 7). Analysis of gene variation among DNA methylated genes by the GO term in the growth process of brindle cattle showed that among the identified genes, the Wnt, MAPK genes associated with melanogenesis, and thyroid hormone related genes were the main cause. Hypomethylation of MAPK and thyroid hormone related genes appeared to occur at a very high rate (Table 3).
Discussion
The difference in the phenotype (heterozygote/homozygote) of the MC1R gene in cattle has been reported to influence the binding of α-MSH (Chung et al., 2000; Seo et al., 2018). This variation also plays an important role in the
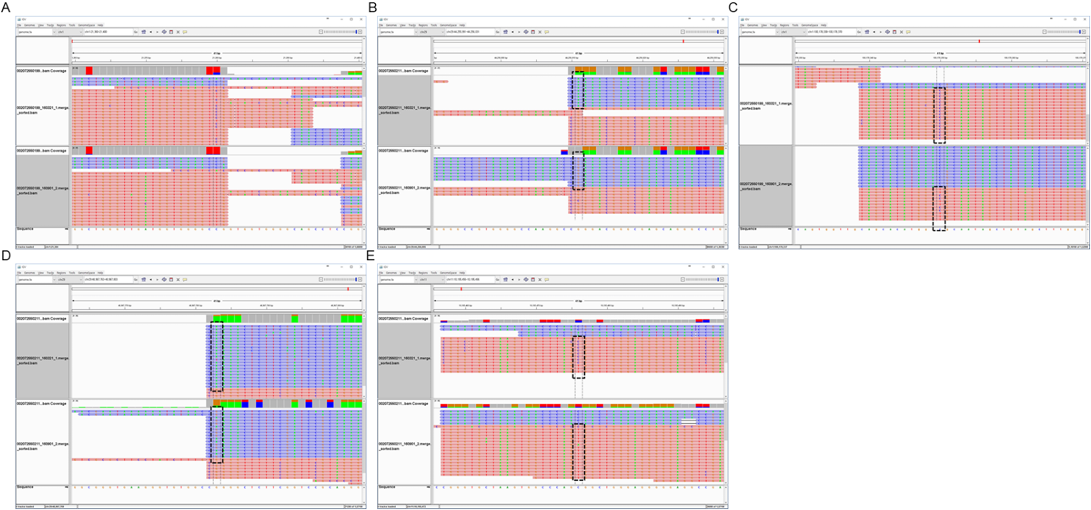
Figure 7: Hypomethylation in the brindle section- A: CpG chr1:21380(+) hypomethylation at brindle 002072550199, B: CpG chr29:44256011(-) hypermethylation at brindle 002072550211, C: CpG chr1:100178350(+) hypomethylation at brindle 002072550318, D: CpG chr11:48967783(-) hypermethylation at brindle 002072550359, E: CpG chr11:10195476(+) hypomethylation at brindle 00207255032.
gene expression of final TYR, TYRP1, and DCT during the process of melanogenesis. However, the variation in MC1R phenotype is either caused by the genetic action or the DNA methylation of the epigenome. Hence, this mechanism of melanogenesis was believed to play a role in the formation of the final metabolite of melanin (Kim et al., 2018). Therefore, this study analyzed whether changes in coat color of brindle cattle and the variation in related genes were characteristically or specially associated with the mechanism of melanogenesis, depending on the type of pigmentation within the melanocytes in brindle cattle (Ishii et al., 2017). Analysis of the expression of genes of the black and yellow color sections of the brindle cattle (brindle color) demonstrated that the expression of major factors such as TYR and DCT, among the genes related to the melanogenesis, in the yellow and brindle sections was more than five times higher than that in the black sections. In other words, this result showed that the action of the melanocytes in the black and yellow sections was different. In this study, as evidence of genetic variation related to melanogenesis, analysis of methylation variation in DNA from blood collected from brindle cattle at the age of less than one month, when the coat color had not formed, and six months, when the coat color had formed, was performed. It demonstrated a 2x75 bp read set of around 80% of the Q30 base of 40 to 70 million reads. Analysis of the gene sequence of CpG formed by Wnt, MARK, and thyroid hormone signaling, which are the major mechanisms of melanogenesis, confirmed that the epigenesis of TYR and MARK-linked genes was very high. In other words, coat color variation in brindle cattle, is believed to be activated as the young cattle grows into the adult phase. The control mechanism is considered to accelerate the expression of coat color. The results of the study by Choi et al. (2013) identified base sequence variations in about 18,000 genes in the results of NGS of brindle and yellow color and suggested the possibility of specific markers for brindle cattle. Additionally, epigenomic variation was found to occur during the mechanisms of melanogenesis, making it possible to develop epigenetic markers. Therefore, the results of this study confirmed that coat color due to melanocyte and keratinocyte pigments in brindle cattle was determined by the different variations in coat color of different species, including dairy.
Conclusion
The classification of breeds according to the phenotype of cattle is a very important factor of securing genetic resources in many countries. But so far, the exact understanding of the phenotype of a cattle’s coat color remains unknown. Therefore, we analyzed the changes in melanogenesis according to the epigenetics of cattle. Our study results proved, the Korean brindle cattle showed an increase in melanogenesis by both MC1R and MARK signaling and the cell cycle active mechanism, resulting in variations in coat color. Additionally, analysis of expression patterns of epigenetic genes of changes in coat color during the growth process of calves in brindle cattle showed that there were changes in the MARK-related and melanogenesis-associated major factors in brindle color formation. The results of this study could be used as important base data for further study of Korean brindle cattle and further demonstrate the unique characteristics of this breed.
Authors Contribution
All authors contributed equally.
Conflict of interest
The authors have declared no conflict of interest.
References